Helios is a key transcriptional regulator of outer hair cell maturation
- PMID: 30464345
- PMCID: PMC6542691
- DOI: 10.1038/s41586-018-0728-4
Helios is a key transcriptional regulator of outer hair cell maturation
Abstract
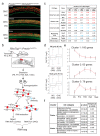
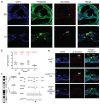
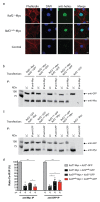
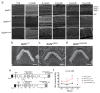
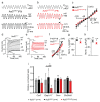
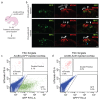
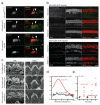
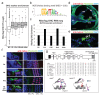
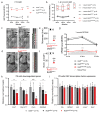
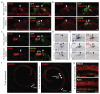
The sensory cells that are responsible for hearing include the cochlear inner hair cells (IHCs) and outer hair cells (OHCs), with the OHCs being necessary for sound sensitivity and tuning1. Both cell types are thought to arise from common progenitors; however, our understanding of the factors that control the fate of IHCs and OHCs remains limited. Here we identify Ikzf2 (which encodes Helios) as an essential transcription factor in mice that is required for OHC functional maturation and hearing. Helios is expressed in postnatal mouse OHCs, and in the cello mouse model a point mutation in Ikzf2 causes early-onset sensorineural hearing loss. Ikzf2cello/cello OHCs have greatly reduced prestin-dependent electromotile activity, a hallmark of OHC functional maturation, and show reduced levels of crucial OHC-expressed genes such as Slc26a5 (which encodes prestin) and Ocm. Moreover, we show that ectopic expression of Ikzf2 in IHCs: induces the expression of OHC-specific genes; reduces the expression of canonical IHC genes; and confers electromotility to IHCs, demonstrating that Ikzf2 can partially shift the IHC transcriptome towards an OHC-like identity.
Conflict of interest statement
The authors declare no competing financial interests.
Figures




References
-
- Ehret G. Development of absolute auditory thresholds in the house mouse (Mus musculus) J Am Audiol Soc. 1976;1:179–84. - PubMed
-
- World Health Organization. Deafness and hearing loss fact sheet. WHO Media centre. 2018 Available at: http://www.who.int/mediacentre/factsheets/fs300/en/
Publication types
MeSH terms
Substances
Grants and funding
- T32 DC000059/DC/NIDCD NIH HHS/United States
- NIH T32DC00046 /DC/NIDCD NIH HHS/United States
- MC_UP_1503/2/MRC_/Medical Research Council/United Kingdom
- R01 DC013817/DC/NIDCD NIH HHS/United States
- U01 DC013817/DC/NIDCD NIH HHS/United States
- MC_U142684175/MRC_/Medical Research Council/United Kingdom
- NIDCD DC000059/DC/NIDCD NIH HHS/United States
- T32 DC000046/DC/NIDCD NIH HHS/United States
- 102892/WT_/Wellcome Trust/United Kingdom
- Z01 DC000059/ImNIH/Intramural NIH HHS/United States
- MC_UP_1502/1/MRC_/Medical Research Council/United Kingdom
- R01 DC003544/DC/NIDCD NIH HHS/United States
- F31 DC016218/DC/NIDCD NIH HHS/United States
- 102892/Z/13/Z/WT_/Wellcome Trust/United Kingdom
- R01 DC014658/DC/NIDCD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

