PANINI: Pangenome Neighbour Identification for Bacterial Populations
- PMID: 30465642
- PMCID: PMC6521588
- DOI: 10.1099/mgen.0.000220
PANINI: Pangenome Neighbour Identification for Bacterial Populations
Abstract
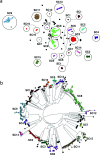
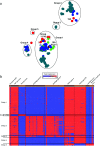
The standard workhorse for genomic analysis of the evolution of bacterial populations is phylogenetic modelling of mutations in the core genome. However, a notable amount of information about evolutionary and transmission processes in diverse populations can be lost unless the accessory genome is also taken into consideration. Here, we introduce panini (Pangenome Neighbour Identification for Bacterial Populations), a computationally scalable method for identifying the neighbours for each isolate in a data set using unsupervised machine learning with stochastic neighbour embedding based on the t-SNE (t-distributed stochastic neighbour embedding) algorithm. panini is browser-based and integrates with the Microreact platform for rapid online visualization and exploration of both core and accessory genome evolutionary signals, together with relevant epidemiological, geographical, temporal and other metadata. Several case studies with single- and multi-clone pneumococcal populations are presented to demonstrate the ability to identify biologically important signals from gene content data. panini is available at http://panini.pathogen.watch and code at http://gitlab.com/cgps/panini.
Keywords: machine learning; microbial population genomics; pangenome; web application.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures