Gut microbiome composition is associated with spatial structuring and social interactions in semi-feral Welsh Mountain ponies
- PMID: 30466491
- PMCID: PMC6251106
- DOI: 10.1186/s40168-018-0593-2
Gut microbiome composition is associated with spatial structuring and social interactions in semi-feral Welsh Mountain ponies
Abstract
Background: Microbiome composition is linked to host functional traits including metabolism and immune function. Drivers of microbiome composition are increasingly well-characterised; however, evidence of group-level microbiome convergence is limited and may represent a multi-level trait (i.e. across individuals and groups), whereby heritable phenotypes are influenced by social interactions. Here, we investigate the influence of spatial structuring and social interactions on the gut microbiome composition of Welsh mountain ponies.
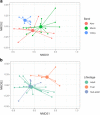
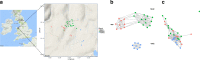
Results: We show that semi-feral ponies exhibit variation in microbiome composition according to band (group) membership, in addition to considerable within-individual variation. Spatial structuring was also identified within bands, suggesting that despite communal living, social behaviours still influence microbiome composition. Indeed, we show that specific interactions (i.e. mother-offspring and stallion-mare) lead to more similar microbiomes, further supporting the notion that individuals influence the microbiome composition of one another and ultimately the group. Foals exhibited different microbiome composition to sub-adults and adults, most likely related to differences in diet.
Conclusions: We provide novel evidence that microbiome composition is structured at multiple levels within populations of social mammals and thus may form a unit on which selection can act. High levels of within-individual variation in microbiome composition, combined with the potential for social interactions to influence microbiome composition, suggest the direction of microbiome selection may be influenced by the individual members present in the group. Although the functional implications of this require further research, these results lend support to the idea that multi-level selection can act on microbiomes.
Keywords: 16S rRNA gene; Amplicon sequencing; Harem; Horizontal transmission; Life stage; Multi-level trait; Social networks; Spatial proximity; Vertical transmission.
Conflict of interest statement
Ethics approval and consent to participate
This study was approved by the University of Salford Research, Innovation, and Academic Engagement Ethical Approval Panel (ST1617-83) and the University of Manchester (Cat-D; non-licenced procedure).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

