Foxi3 transcription factor activity is mediated by a C-terminal transactivation domain and regulated by the Protein Phosphatase 2A (PP2A) complex
- PMID: 30467319
- PMCID: PMC6250667
- DOI: 10.1038/s41598-018-35390-8
Foxi3 transcription factor activity is mediated by a C-terminal transactivation domain and regulated by the Protein Phosphatase 2A (PP2A) complex
Abstract
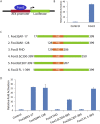
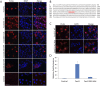
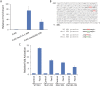
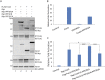
The Forkhead box (FOX) family consists of at least 19 subgroups of transcription factors which are characterized by the presence of an evolutionary conserved 'forkhead' or 'winged-helix' DNA-binding domain. Despite having a conserved core DNA binding domain, FOX proteins display remarkable functional diversity and are involved in many developmental and cell specific processes. In the present study, we focus on a poorly characterized member of the Forkhead family, Foxi3, which plays a critical role in the development of the inner ear and jaw. We show that Foxi3 contains at least two important functional domains, a nuclear localization sequence (NLS) and a C-terminal transactivation domain (TAD), and that it directly binds its targets in a sequence specific manner. We also show that the transcriptional activity of Foxi3 is regulated by phosphorylation, and that the activity of Foxi3 can be attenuated by its physical interaction with the protein phosphatase 2A (PP2A) complex.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Defining the Protein Phosphatase 2A (PP2A) Subcomplexes That Regulate FoxO Transcription Factor Localization.Cells. 2025 Feb 27;14(5):342. doi: 10.3390/cells14050342. Cells. 2025. PMID: 40072071 Free PMC article.
-
Protein phosphatase 2A (B55α) prevents premature activation of forkhead transcription factor FoxM1 by antagonizing cyclin A/cyclin-dependent kinase-mediated phosphorylation.J Biol Chem. 2011 Sep 23;286(38):33029-36. doi: 10.1074/jbc.M111.253724. Epub 2011 Aug 3. J Biol Chem. 2011. PMID: 21813648 Free PMC article.
-
Multiple modes of chromatin remodeling by Forkhead box proteins.Biochim Biophys Acta. 2012 Jul;1819(7):707-15. doi: 10.1016/j.bbagrm.2012.02.018. Epub 2012 Mar 2. Biochim Biophys Acta. 2012. PMID: 22406422 Review.
-
A conserved cation binding site in the DNA binding domain of forkhead box transcription factors regulates DNA binding by FOXP2.Arch Biochem Biophys. 2018 Nov 1;657:56-64. doi: 10.1016/j.abb.2018.09.009. Epub 2018 Sep 15. Arch Biochem Biophys. 2018. PMID: 30227110
-
Forkhead Transcription Factors in Health and Disease.Trends Genet. 2021 May;37(5):460-475. doi: 10.1016/j.tig.2020.11.003. Epub 2020 Dec 7. Trends Genet. 2021. PMID: 33303287 Review.
Cited by
-
Damaging variants in FOXI3 cause microtia and craniofacial microsomia.Genet Med. 2023 Jan;25(1):143-150. doi: 10.1016/j.gim.2022.09.005. Epub 2022 Oct 19. Genet Med. 2023. PMID: 36260083 Free PMC article.
-
Characterization of novel, recurrent genomic rearrangements as sensitive MRD targets in childhood B-cell precursor ALL.Blood Cancer J. 2019 Nov 29;9(12):96. doi: 10.1038/s41408-019-0257-x. Blood Cancer J. 2019. PMID: 31784504 Free PMC article.
-
FOXI3 pathogenic variants cause one form of craniofacial microsomia.Nat Commun. 2023 Apr 11;14(1):2026. doi: 10.1038/s41467-023-37703-6. Nat Commun. 2023. PMID: 37041148 Free PMC article.
-
Identification of potential molecular mechanism related to craniofacial dysmorphism caused by FOXI3 deficiency.Mol Genet Genomic Med. 2024 Mar;12(3):e2411. doi: 10.1002/mgg3.2411. Mol Genet Genomic Med. 2024. PMID: 38433559 Free PMC article.
-
The Foxi3 transcription factor is necessary for the fate restriction of placodal lineages at the neural plate border.Development. 2023 Oct 1;150(19):dev202047. doi: 10.1242/dev.202047. Epub 2023 Oct 9. Development. 2023. PMID: 37756587 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

