Hotspots for Initiation of Meiotic Recombination
- PMID: 30467513
- PMCID: PMC6237102
- DOI: 10.3389/fgene.2018.00521
Hotspots for Initiation of Meiotic Recombination
Abstract
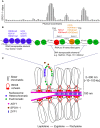
Homologous chromosomes must pair and recombine to ensure faithful chromosome segregation during meiosis, a specialized type of cell division that occurs in sexually reproducing eukaryotes. Meiotic recombination initiates by programmed induction of DNA double-strand breaks (DSBs) by the conserved type II topoisomerase-like enzyme SPO11. A subset of meiotic DSBs are resolved as crossovers, whereby reciprocal exchange of DNA occurs between homologous chromosomes. Importantly, DSBs are non-randomly distributed along eukaryotic chromosomes, forming preferentially in permissive regions known as hotspots. In many species, including plants, DSB hotspots are located within nucleosome-depleted regions. DSB localization is governed by interconnected factors, including cis-regulatory elements, transcription factor binding, and chromatin accessibility, as well as by higher-order chromosome architecture. The spatiotemporal control of DSB formation occurs within a specialized chromosomal structure characterized by sister chromatids organized into linear arrays of chromatin loops that are anchored to a proteinaceous axis. Although SPO11 and its partner proteins required for DSB formation are bound to the axis, DSBs occur preferentially within the chromatin loops, which supports the "tethered-loop/axis model" for meiotic recombination. In this mini review, we discuss insights gained from recent efforts to define and profile DSB hotspots at high resolution in eukaryotic genomes. These advances are deepening our understanding of how meiotic recombination shapes genetic diversity and genome evolution in diverse species.
Keywords: DSB; chromatin; crossover; epigenetics; hotspot; meiosis; nucleosomes; recombination.
Figures
Similar articles
-
Nucleosomes and DNA methylation shape meiotic DSB frequency in Arabidopsis thaliana transposons and gene regulatory regions.Genome Res. 2018 Apr;28(4):532-546. doi: 10.1101/gr.225599.117. Epub 2018 Mar 12. Genome Res. 2018. PMID: 29530928 Free PMC article.
-
High-Resolution Global Analysis of the Influences of Bas1 and Ino4 Transcription Factors on Meiotic DNA Break Distributions in Saccharomyces cerevisiae.Genetics. 2015 Oct;201(2):525-42. doi: 10.1534/genetics.115.178293. Epub 2015 Aug 5. Genetics. 2015. PMID: 26245832 Free PMC article.
-
The conserved histone variant H2A.Z illuminates meiotic recombination initiation.Curr Genet. 2018 Oct;64(5):1015-1019. doi: 10.1007/s00294-018-0825-9. Epub 2018 Mar 16. Curr Genet. 2018. PMID: 29549582 Review.
-
Spatiotemporal regulation of meiotic recombination by Liaisonin.Bioarchitecture. 2013 Jan-Feb;3(1):20-4. doi: 10.4161/bioa.23966. Bioarchitecture. 2013. PMID: 23572041 Free PMC article.
-
Advances Towards How Meiotic Recombination Is Initiated: A Comparative View and Perspectives for Plant Meiosis Research.Int J Mol Sci. 2019 Sep 23;20(19):4718. doi: 10.3390/ijms20194718. Int J Mol Sci. 2019. PMID: 31547623 Free PMC article. Review.
Cited by
-
DNA polymerase epsilon interacts with SUVH2/9 to repress the expression of genes associated with meiotic DSB hotspot in Arabidopsis.Proc Natl Acad Sci U S A. 2022 Oct 11;119(41):e2208441119. doi: 10.1073/pnas.2208441119. Epub 2022 Oct 3. Proc Natl Acad Sci U S A. 2022. PMID: 36191225 Free PMC article.
-
Diverse DNA Sequence Motifs Activate Meiotic Recombination Hotspots Through a Common Chromatin Remodeling Pathway.Genetics. 2019 Nov;213(3):789-803. doi: 10.1534/genetics.119.302679. Epub 2019 Sep 11. Genetics. 2019. PMID: 31511300 Free PMC article.
-
Meiotic Genes and DNA Double Strand Break Repair in Cancer.Front Genet. 2022 Feb 18;13:831620. doi: 10.3389/fgene.2022.831620. eCollection 2022. Front Genet. 2022. PMID: 35251135 Free PMC article. Review.
-
Manipulation of Meiotic Recombination to Hasten Crop Improvement.Biology (Basel). 2022 Feb 25;11(3):369. doi: 10.3390/biology11030369. Biology (Basel). 2022. PMID: 35336743 Free PMC article. Review.
-
dCas9-SPO11-1 locally stimulates meiotic recombination in rice.Front Plant Sci. 2025 May 1;16:1580225. doi: 10.3389/fpls.2025.1580225. eCollection 2025. Front Plant Sci. 2025. PMID: 40376157 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources

