The sequence specificity of homeodomain-DNA interaction
- PMID: 3046753
- PMCID: PMC2753412
- DOI: 10.1016/0092-8674(88)90123-7
The sequence specificity of homeodomain-DNA interaction
Abstract
The Drosophila developmental gene, engrailed, encodes a sequence-specific DNA binding activity. Using deletion constructs expressed as fusion proteins in E. coli, we localized this activity to the conserved homeodomain (HD). The binding site consensus, TCAATTAAAT, is found in clusters in the engrailed regulatory region. Weak binding of the En HD to one copy of a synthetic consensus is enhanced by adjacent copies. The distantly related HD encoded by fushi tarazu binds to the same sites as the En HD, but differs in its preference for related sites. Both HDs bind a second type of sequence, a repeat of TAA. The similarity in sequence specificity of En and Ftz HDs suggests that, within families of DNA binding proteins, close relatives will exhibit similar specificities. Competition among related regulatory proteins might govern which protein occupies a given binding site and consequently determine the ultimate effect of cis-acting regulatory sites.
Figures

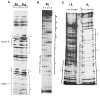


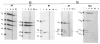
References
-
- Arriza JL, Weinberger C, Cerelli G, Glaser TM, Handelin BL, Housman DE, Evans RM. Cloning of human mineralocorticoid receptor complimentary DNA: structural and functional kinship with the glucocorticoid receptor. Science. 1987;237:268–274. - PubMed
-
- Beachy PA, Helfand SL, Hogness DS. Segmental distribution of bithorax complex proteins during Drosophila development. Nature. 1985;313:545–551. - PubMed
-
- Bopp D, Burri M, Baumgartner S, Frigerio G, Noll M. Conservation of a large protein domain in the segmentation gene paired and in functionally related genes in Drosophila. Cell. 1986;47:1033–1049. - PubMed
-
- Carroll SB, Scott MP. Localization of the fushi tarazu protein during Drosophila embryogenesis. Cell. 1985;43:47–57. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

