Millisecond Timescale Motions Connect Amino Acid Interaction Networks in Alpha Tryptophan Synthase
- PMID: 30467546
- PMCID: PMC6236060
- DOI: 10.3389/fmolb.2018.00092
Millisecond Timescale Motions Connect Amino Acid Interaction Networks in Alpha Tryptophan Synthase
Abstract
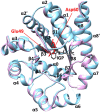
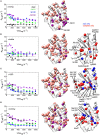
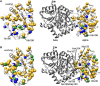
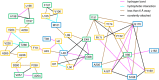
Tryptophan synthase is a model system for understanding allosteric regulation within enzyme complexes. Amino acid interaction networks were previously delineated in the isolated alpha subunit (αTS) in the absence of the beta subunit (βTS). The amino acid interaction networks were different between the ligand-free enzyme and the enzyme actively catalyzing turnover. Previous X-ray crystallography studies indicated only minor localized changes when ligands bind αTS, and so, structural changes alone could not explain the changes to the amino acid interaction networks. We hypothesized that the network changes could instead be related to changes in conformational dynamics. As such, we conducted nuclear magnetic resonance relaxation studies on different substrate- and products-bound complexes of αTS. Specifically, we collected 15N R2 relaxation dispersion data that reports on microsecond-to-millisecond timescale motion of backbone amide groups. These experiments indicated that there are conformational exchange events throughout αTS. Substrate and product binding change specific motional pathways throughout the enzyme, and these pathways connect the previously identified network residues. These pathways reach the αTS/βTS binding interface, suggesting that the identified dynamic networks may also be important for communication with the βTS subunit.
Keywords: allostery; amino acid networks; enzyme dynamics; enzyme regulation; protein NMR; relaxation dispersion; tryptophan synthase.
Figures
References
LinkOut - more resources
Full Text Sources
Research Materials

