Fluorescent peptide displacement as a general assay for screening small molecule libraries against RNA
- PMID: 30468226
- PMCID: PMC6503966
- DOI: 10.1039/c8ob02467g
Fluorescent peptide displacement as a general assay for screening small molecule libraries against RNA
Abstract
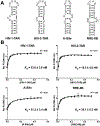
A prominent hurdle in developing small molecule probes against RNA is the relative scarcity of general screening methods. In this study, we demonstrate the application of a fluorescent peptide displacement assay to screen small molecule probes against four different RNA targets. The designed experimental protocol combined with statistical analysis provides a fast and convenient method to simultaneously evaluate small molecule libraries against different RNA targets and classify them based on affinity and selectivity patterns.
Conflict of interest statement
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

