FMRP Interacts with C/D Box snoRNA in the Nucleus and Regulates Ribosomal RNA Methylation
- PMID: 30469012
- PMCID: PMC6249352
- DOI: 10.1016/j.isci.2018.11.007
FMRP Interacts with C/D Box snoRNA in the Nucleus and Regulates Ribosomal RNA Methylation
Erratum in
-
FMRP Interacts with C/D Box snoRNA in the Nucleus and Regulates Ribosomal RNA Methylation.iScience. 2019 Feb 22;12:368. doi: 10.1016/j.isci.2019.01.026. Epub 2019 Feb 11. iScience. 2019. PMID: 30763793 Free PMC article. No abstract available.
Abstract
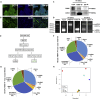
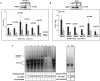
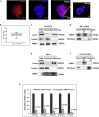
FMRP is an RNA-binding protein that is known to localize in the cytoplasm and in the nucleus. Here, we have identified an interaction of FMRP with a specific set of C/D box snoRNAs in the nucleus. C/D box snoRNAs guide 2'O methylations of ribosomal RNA (rRNA) on defined sites, and this modification regulates rRNA folding and assembly of ribosomes. 2'O methylation of rRNA is partial on several sites in human embryonic stem cells, which results in ribosomes with differential methylation patterns. FMRP-snoRNA interaction affects rRNA methylation on several of these sites, and in the absence of FMRP, differential methylation pattern of rRNA is significantly altered. We found that FMRP recognizes ribosomes carrying specific methylation patterns on rRNA and the recognition of methylation pattern by FMRP may potentially determine the translation status of its target mRNAs. Thus, FMRP integrates its function in the nucleus and in the cytoplasm.
Keywords: Molecular Interaction; Omics; Stem Cells Research.
Copyright © 2018 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures




References
-
- Bardoni B., Willemsen R., Weiler I.J., Schenck A., Severijnen L.A., Hindelang C., Lalli E., Mandel J.L. NUFIP1 (nuclear FMRP interacting protein 1) is a nucleocytoplasmic shuttling protein associated with active synaptoneurosomes. Exp. Cell Res. 2003;289:95–107. - PubMed
-
- Bensaddek D., Nicolas A., Lamond A.I. Quantitative proteomic analysis of the human nucleolus. Methods Mol. Biol. 2016;1455:249–262. - PubMed
-
- Cavaille J., Nicoloso M., Bachellerie J.P. Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature. 1996;383:732–735. - PubMed
LinkOut - more resources
Full Text Sources

