Oncogenic KRAS supports pancreatic cancer through regulation of nucleotide synthesis
- PMID: 30470748
- PMCID: PMC6251888
- DOI: 10.1038/s41467-018-07472-8
Oncogenic KRAS supports pancreatic cancer through regulation of nucleotide synthesis
Abstract
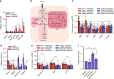
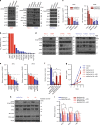
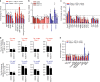
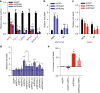
Oncogenic KRAS is the key driver of pancreatic ductal adenocarcinoma (PDAC). We previously described a role for KRAS in PDAC tumor maintenance through rewiring of cellular metabolism to support proliferation. Understanding the details of this metabolic reprogramming in human PDAC may provide novel therapeutic opportunities. Here we show that the dependence on oncogenic KRAS correlates with specific metabolic profiles that involve maintenance of nucleotide pools as key mediators of KRAS-dependence. KRAS promotes these effects by activating a MAPK-dependent signaling pathway leading to MYC upregulation and transcription of the non-oxidative pentose phosphate pathway (PPP) gene RPIA, which results in nucleotide biosynthesis. The use of MEK inhibitors recapitulates the KRAS-dependence pattern and the expected metabolic changes. Antagonizing the PPP or pyrimidine biosynthesis inhibits the growth of KRAS-resistant cells. Together, these data reveal differential metabolic rewiring between KRAS-resistant and sensitive cells, and demonstrate that targeting nucleotide metabolism can overcome resistance to KRAS/MEK inhibition.
Conflict of interest statement
A.C.K. has financial interests in Vescor Therapeutics, LLC. A.C.K. is an inventor on patents pertaining to KRAS regulated metabolic pathways, redox control pathways in pancreatic cancer, targeting GOT1 as a therapeutic approach, and the autophagic control of iron metabolism. A.C.K. is on the SAB of Cornerstone/Rafael Pharmaceuticals.
Figures


References
-
- National Cancer Institute. M. SEER Cancer Statistics Factsheets: Pancreas Cancer. (National Cancer Institute, Bethesda, 2012).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

