Transcription factor Ptf1a in development, diseases and reprogramming
- PMID: 30470852
- PMCID: PMC11105224
- DOI: 10.1007/s00018-018-2972-z
Transcription factor Ptf1a in development, diseases and reprogramming
Abstract
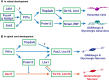
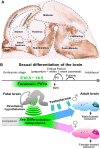
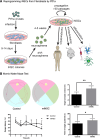
The transcription factor Ptf1a is a crucial helix-loop-helix (bHLH) protein selectively expressed in the pancreas, retina, spinal cord, brain, and enteric nervous system. Ptf1a is preferably assembled into a transcription trimeric complex PTF1 with an E protein and Rbpj (or Rbpjl). In pancreatic development, Ptf1a is indispensable in controlling the expansion of multipotent progenitor cells as well as the specification and maintenance of the acinar cells. In neural tissues, Ptf1a is transiently expressed in the post-mitotic cells and specifies the inhibitory neuronal cell fates, mostly mediated by downstream genes such as Tfap2a/b and Prdm13. Mutations in the coding and non-coding regulatory sequences resulting in Ptf1a gain- or loss-of-function are associated with genetic diseases such as pancreatic and cerebellar agenesis in the rodent and human. Surprisingly, Ptf1a alone is sufficient to reprogram mouse or human fibroblasts into tripotential neural stem cells. Its pleiotropic functions in many biological processes remain to be deciphered in the future.
Keywords: Acinar cells; Cell fate specification; Diabetes; GABAergic; Glutamatergic; Glycinergic; Inheritable; Inhibitory neurotransmitter; Pancreatic development; Retinal development; Somatic cell reprogramming; Transcriptional regulation.
Figures




Similar articles
-
Early pancreatic development requires the vertebrate Suppressor of Hairless (RBPJ) in the PTF1 bHLH complex.Genes Dev. 2007 Oct 15;21(20):2629-43. doi: 10.1101/gad.1575207. Genes Dev. 2007. PMID: 17938243 Free PMC article.
-
Neurog2 is a direct downstream target of the Ptf1a-Rbpj transcription complex in dorsal spinal cord.Development. 2009 Sep;136(17):2945-54. doi: 10.1242/dev.035352. Epub 2009 Jul 29. Development. 2009. PMID: 19641016 Free PMC article.
-
The Prdm13 histone methyltransferase encoding gene is a Ptf1a-Rbpj downstream target that suppresses glutamatergic and promotes GABAergic neuronal fate in the dorsal neural tube.Dev Biol. 2014 Feb 15;386(2):340-57. doi: 10.1016/j.ydbio.2013.12.024. Epub 2013 Dec 24. Dev Biol. 2014. PMID: 24370451
-
bHLH transcription factors in neural development, disease, and reprogramming.Brain Res. 2019 Feb 15;1705:48-65. doi: 10.1016/j.brainres.2018.03.013. Epub 2018 Mar 12. Brain Res. 2019. PMID: 29544733 Review.
-
Ptf1a function and transcriptional cis-regulation, a cornerstone in vertebrate pancreas development.FEBS J. 2022 Sep;289(17):5121-5136. doi: 10.1111/febs.16075. Epub 2021 Jun 30. FEBS J. 2022. PMID: 34125483 Free PMC article. Review.
Cited by
-
Notch pathway mutants do not equivalently perturb mouse embryonic retinal development.PLoS Genet. 2023 Sep 26;19(9):e1010928. doi: 10.1371/journal.pgen.1010928. eCollection 2023 Sep. PLoS Genet. 2023. PMID: 37751417 Free PMC article.
-
The dorsal/ventral subdivision of the hindbrain predates the tunicate/vertebrate split.bioRxiv [Preprint]. 2025 Jul 18:2025.07.15.664975. doi: 10.1101/2025.07.15.664975. bioRxiv. 2025. PMID: 40791562 Free PMC article. Preprint.
-
Selective deletion of zinc transporter 3 in amacrine cells promotes retinal ganglion cell survival and optic nerve regeneration after injury.Neural Regen Res. 2023 Dec;18(12):2773-2780. doi: 10.4103/1673-5374.373660. Neural Regen Res. 2023. PMID: 37449644 Free PMC article.
-
Promoter hypermethylation of neural-related genes is compatible with stemness in solid cancers.Epigenetics Chromatin. 2023 Aug 3;16(1):31. doi: 10.1186/s13072-023-00505-7. Epigenetics Chromatin. 2023. PMID: 37537688 Free PMC article.
-
Ptf1a expression is necessary for correct targeting of spiral ganglion neurons within the cochlear nuclei.Neurosci Lett. 2023 May 29;806:137244. doi: 10.1016/j.neulet.2023.137244. Epub 2023 Apr 11. Neurosci Lett. 2023. PMID: 37055006 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

