Analysis of spliceosome dynamics by maximum likelihood fitting of dwell time distributions
- PMID: 30472247
- PMCID: PMC6363122
- DOI: 10.1016/j.ymeth.2018.11.014
Analysis of spliceosome dynamics by maximum likelihood fitting of dwell time distributions
Abstract
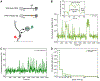
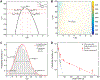
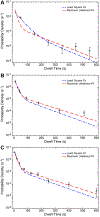
Colocalization single-molecule methods can provide a wealth of information concerning the ordering and dynamics of biomolecule assembly. These have been used extensively to study the pathways of spliceosome assembly in vitro. Key to these experiments is the measurement of binding times-either the dwell times of a multi-molecular interaction or times in between binding events. By analyzing hundreds of these times, many new insights into the kinetic pathways governing spliceosome assembly have been obtained. Collections of binding times are often plotted as histograms and can be fit to kinetic models using a variety of methods. Here, we describe the use of maximum likelihood methods to fit dwell time distributions without binning. In addition, we discuss several aspects of analyzing these distributions with histograms and pitfalls that can be encountered if improperly binned histograms are used. We have automated several aspects of maximum likelihood fitting of dwell time distributions in the AGATHA software package.
Keywords: Dynamics; Fitting; Fluorescence; Single-molecule; Software; Spliceosome.
Copyright © 2018 Elsevier Inc. All rights reserved.
Figures

References
-
- Nilsen TW, The spliceosome: the most complex macromolecular machine in the cell?, Bioessays 25(12) (2003) 1147–1149. - PubMed
-
- Wahl MC, Will CL, Lührmann R, The spliceosome: design principles of a dynamic RNP machine, Cell 136(4) (2009) 701–718. - PubMed
-
- Shi Y, Mechanistic insights into precursor messenger RNA splicing by the spliceosome, Nature Reviews Molecular Cell Biology 18(11) (2017) 655. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

