Evolutionary biology of human hepatitis viruses
- PMID: 30472320
- PMCID: PMC7114834
- DOI: 10.1016/j.jhep.2018.11.010
Evolutionary biology of human hepatitis viruses
Abstract
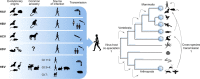
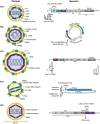
Hepatitis viruses are major threats to human health. During the last decade, highly diverse viruses related to human hepatitis viruses were found in animals other than primates. Herein, we describe both surprising conservation and striking differences of the unique biological properties and infection patterns of human hepatitis viruses and their animal homologues, including transmission routes, liver tropism, oncogenesis, chronicity, pathogenesis and envelopment. We discuss the potential for translation of newly discovered hepatitis viruses into preclinical animal models for drug testing, studies on pathogenesis and vaccine development. Finally, we re-evaluate the evolutionary origins of human hepatitis viruses and discuss the past and present zoonotic potential of their animal homologues.
Keywords: Evolution; Hepatitis; Homologues; Virus; Zoonotic.
Copyright © 2018 European Association for the Study of the Liver. Published by Elsevier B.V. All rights reserved.
Figures





References
-
- (WHO) WHO. Global hepatitis report, 2017. https://www.who.int/hepatitis/publications/global-hepatitis-report2017/en/
-
- (WHO) WHO. Hepatitis C fact sheet. 2017; http://www.who.int/news-room/fact-sheets/detail/hepatitis-c.
-
- Romeo R., Del Ninno E., Rumi M., Russo A., Sangiovanni A., de Franchis R. A 28-year study of the course of hepatitis Delta infection: a risk factor for cirrhosis and hepatocellular carcinoma. Gastroenterology. 2009;136:1629–1638. - PubMed
-
- Krugman S., Giles J.P., Hammond J. Infectious hepatitis. Evidence for two distinctive clinical, epidemiological, and immunological types of infection. JAMA. 1967;200:365–373. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

