Ribosome Stoichiometry: From Form to Function
- PMID: 30473427
- PMCID: PMC6340777
- DOI: 10.1016/j.tibs.2018.10.009
Ribosome Stoichiometry: From Form to Function
Abstract
The existence of eukaryotic ribosomes with distinct ribosomal protein (RP) stoichiometry and regulatory roles in protein synthesis has been speculated for over 60 years. Recent advances in mass spectrometry (MS) and high-throughput analysis have begun to identify and characterize distinct ribosome stoichiometry in yeast and mammalian systems. In addition to RP stoichiometry, ribosomes host a vast array of protein modifications, effectively expanding the number of human RPs from 80 to many thousands of distinct proteoforms. Is it possible that these proteoforms combine to function as a 'ribosome code' to tune protein synthesis? We outline the specific benefits that translational regulation by specialized ribosomes can offer and discuss the means and methodologies available to correlate and characterize RP stoichiometry with function. We highlight previous research with a focus on formulating hypotheses that can guide future experiments and crack the ribosome code.
Keywords: heterogeneity; mass spectrometry; ribosome; stoichiometry; translation.
Copyright © 2018 Elsevier Ltd. All rights reserved.
Figures

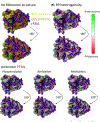

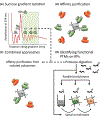
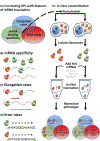
References
-
- Crick FH (1958) On protein synthesis. Symp. Soc. Exp. Biol 12, 138–63 - PubMed
-
- Lodish HF (1974) Model for the regulation of mRNA translation applied to haemoglobin synthesis. Nature 251, 385–8 - PubMed
-
- Mills EW and Green R (2017) Ribosomopathies: There’s strength in numbers. Science 358, eaan2755 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

