Fermentative Production of N-Methylglutamate From Glycerol by Recombinant Pseudomonas putida
- PMID: 30474025
- PMCID: PMC6237917
- DOI: 10.3389/fbioe.2018.00159
Fermentative Production of N-Methylglutamate From Glycerol by Recombinant Pseudomonas putida
Abstract
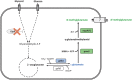
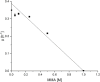
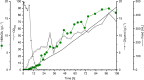
N-methylated amino acids are present in diverse biological molecules in bacteria, archaea and eukaryotes. There is an increasing interest in this molecular class of alkylated amino acids by the pharmaceutical and chemical industries. N-alkylated amino acids have desired functions such as higher proteolytic stability, enhanced membrane permeability and longer peptide half-lives, which are important for the peptide-based drugs, the so-called peptidomimetics. Chemical synthesis of N-methylated amino acids often is limited by incomplete stereoselectivity, over-alkylation or the use of hazardous chemicals. Here, we describe metabolic engineering of Pseudomonas putida KT2440 for the fermentative production of N-methylglutamate from simple carbon sources and monomethylamine. P. putida KT2440, which is generally recognized as safe and grows with glucose and the alternative feedstock glycerol as sole carbon and energy source, was engineered for the production of N-methylglutamate using heterologous enzymes from Methylobacterium extorquens. About 3.9 g L-1 N-methylglutamate accumulated within 48 h in shake flask cultures with minimal medium containing monomethylamine and glycerol. A fed-batch cultivation process yielded a N-methylglutamate titer of 17.9 g L-1.
Keywords: GS/GOGAT; GlpR; Methylobacterium extorquens; N-methylglutamate; N-methylglutamate synthase; Pseudomonas putida; fed-batch fermentation; γ-glutamylmethylamide synthetase.
Figures

Similar articles
-
Fermentative Production of N-Alkylated Glycine Derivatives by Recombinant Corynebacterium glutamicum Using a Mutant of Imine Reductase DpkA From Pseudomonas putida.Front Bioeng Biotechnol. 2019 Sep 26;7:232. doi: 10.3389/fbioe.2019.00232. eCollection 2019. Front Bioeng Biotechnol. 2019. PMID: 31616665 Free PMC article.
-
Metabolic Engineering of Pseudomonas putida for Fermentative Production of l-Theanine.J Agric Food Chem. 2021 Sep 1;69(34):9849-9858. doi: 10.1021/acs.jafc.1c03240. Epub 2021 Aug 17. J Agric Food Chem. 2021. PMID: 34465093
-
γ-Glutamylation of Isopropylamine by Fermentation.Chembiochem. 2024 Jan 15;25(2):e202300608. doi: 10.1002/cbic.202300608. Epub 2023 Nov 29. Chembiochem. 2024. PMID: 37987374
-
Metabolic engineering of Pseudomonas putida KT2440 for high-yield production of protocatechuic acid.Bioresour Technol. 2021 Jan;319:124239. doi: 10.1016/j.biortech.2020.124239. Epub 2020 Oct 7. Bioresour Technol. 2021. PMID: 33254462
-
Microbial Engineering for Production of N-Functionalized Amino Acids and Amines.Biotechnol J. 2020 Jul;15(7):e1900451. doi: 10.1002/biot.201900451. Epub 2020 Mar 30. Biotechnol J. 2020. PMID: 32170807 Review.
Cited by
-
Pseudomonas putida KT2440 is HV1 certified, not GRAS.Microb Biotechnol. 2019 Sep;12(5):845-848. doi: 10.1111/1751-7915.13443. Epub 2019 Jun 14. Microb Biotechnol. 2019. PMID: 31199068 Free PMC article.
-
Sustainable Production of N-methylphenylalanine by Reductive Methylamination of Phenylpyruvate Using Engineered Corynebacterium glutamicum.Microorganisms. 2021 Apr 13;9(4):824. doi: 10.3390/microorganisms9040824. Microorganisms. 2021. PMID: 33924554 Free PMC article.
-
Improved Plasmid-Based Inducible and Constitutive Gene Expression in Corynebacterium glutamicum.Microorganisms. 2021 Jan 19;9(1):204. doi: 10.3390/microorganisms9010204. Microorganisms. 2021. PMID: 33478126 Free PMC article.
-
Prospects of formamide as nitrogen source in biotechnological production processes.Appl Microbiol Biotechnol. 2024 Dec;108(1):105. doi: 10.1007/s00253-023-12962-x. Epub 2024 Jan 10. Appl Microbiol Biotechnol. 2024. PMID: 38204134 Free PMC article. Review.
-
Fermentative Production of N-Alkylated Glycine Derivatives by Recombinant Corynebacterium glutamicum Using a Mutant of Imine Reductase DpkA From Pseudomonas putida.Front Bioeng Biotechnol. 2019 Sep 26;7:232. doi: 10.3389/fbioe.2019.00232. eCollection 2019. Front Bioeng Biotechnol. 2019. PMID: 31616665 Free PMC article.
References
-
- Bagdasarian M., Lurz R., Rückert B., Franklin F. C., Bagdasarian M. M., Frey J., et al. . (1981). Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene 16, 237–247. 10.1016/0378-1119(81)90080-9 - DOI - PubMed
LinkOut - more resources
Full Text Sources

