Telomere elongation through hTERT immortalization leads to chromosome repositioning in control cells and genomic instability in Hutchinson-Gilford progeria syndrome fibroblasts, expressing a novel SUN1 isoform
- PMID: 30474255
- PMCID: PMC6590296
- DOI: 10.1002/gcc.22711
Telomere elongation through hTERT immortalization leads to chromosome repositioning in control cells and genomic instability in Hutchinson-Gilford progeria syndrome fibroblasts, expressing a novel SUN1 isoform
Abstract
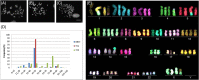
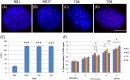
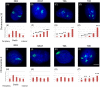
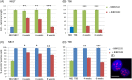
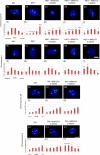
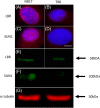
Immortalizing primary cells with human telomerase reverse transcriptase (hTERT) has been common practice to enable primary cells to be of extended use in the laboratory because they avoid replicative senescence. Studying exogenously expressed hTERT in cells also affords scientists models of early carcinogenesis and telomere behavior. Control and the premature ageing disease-Hutchinson-Gilford progeria syndrome (HGPS) primary dermal fibroblasts, with and without the classical G608G mutation have been immortalized with exogenous hTERT. However, hTERT immortalization surprisingly elicits genome reorganization not only in disease cells but also in the normal control cells, such that whole chromosome territories normally located at the nuclear periphery in proliferating fibroblasts become mislocalized in the nuclear interior. This includes chromosome 18 in the control fibroblasts and both chromosomes 18 and X in HGPS cells, which physically express an isoform of the LINC complex protein SUN1 that has previously only been theoretical. Additionally, this HGPS cell line has also become genomically unstable and has a tetraploid karyotype, which could be due to the novel SUN1 isoform. Long-term treatment with the hTERT inhibitor BIBR1532 enabled the reduction of telomere length in the immortalized cells and resulted that these mislocalized internal chromosomes to be located at the nuclear periphery, as assessed in actively proliferating cells. Taken together, these findings reveal that elongated telomeres lead to dramatic chromosome mislocalization, which can be restored with a drug treatment that results in telomere reshortening and that a novel SUN1 isoform combined with elongated telomeres leads to genomic instability. Thus, care should be taken when interpreting data from genomic studies in hTERT-immortalized cell lines.
Keywords: BIBR1532; Hutchinson-Gilford progeria syndrome; M-FISH; Q-FISH; SUN1; SUN1 isoform 9; chromosome territories; genomic instability; hTERT; telomeres.
© 2018 The Authors. Genes, Chromosomes & Cancer published by Wiley Periodicals, Inc.
Figures

References
-
- Campisi J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbours. Cell. 2005;120(4):513‐522. - PubMed
-
- Blackburn EH. Telomeres and telomerase: their mechanisms of action and the effects of altering their functions. FEBS Lett. 2005;579(4):859‐862. - PubMed
-
- Blackburn EH. Telomere states and cell fates. Nature. 2000;408(6808):53‐56. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

