The role of DNA methylation in human trophoblast differentiation
- PMID: 30475094
- PMCID: PMC6986789
- DOI: 10.1080/15592294.2018.1549462
The role of DNA methylation in human trophoblast differentiation
Abstract
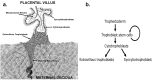
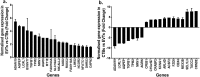
The placenta is a vital fetal exchange organ connecting mother and baby. Specialised placental epithelial cells, called trophoblasts, are essential for adequate placental function. Trophoblasts transform the maternal vasculature to allow efficient blood flow to the placenta and facilitate adequate nutrient uptake. Placental development is in part regulated by epigenetic mechanisms. However, our understanding of how DNA methylation contributes to human trophoblast differentiation is limited. To better understand how genome-wide methylation differences affect trophoblast differentiation, reduced representation bisulfite sequencing (RRBS) was conducted on four matched sets of trophoblasts; side-population trophoblasts (a candidate human trophoblast stem cell population), cytotrophoblasts (an intermediate progenitor population), and extravillous trophoblasts (EVT, a terminally differentiated population) each isolated from the same first trimester placenta. Each trophoblast population had a distinct methylome. In line with their close differentiation relationship, the methylation profile of side-population trophoblasts was most similar to cytotrophoblasts, whilst EVT had the most distinct methylome. In comparison to mature trophoblast populations, side-population trophoblasts exhibited differential methylation of genes and miRNAs involved in cell cycle regulation, differentiation, and regulation of pluripotency. A combined methylomic and transcriptomic approach was taken to better understand cytotrophoblast differentiation to EVT. This revealed methylation of 41 genes involved in epithelial to mesenchymal transition and metastatic cancer pathways, which likely contributes to the acquisition of an invasive EVT phenotype. However, the methylation status of a gene did not always predict gene expression. Therefore, while CpG methylation plays a role in trophoblast differentiation, it is likely not the only regulatory mechanism involved in this process.
Keywords: DNA methylation; Placenta; cytotrophoblasts; extravillous trophoblasts; trophoblast.
Figures




References
-
- Benirschke K. The placenta: structure and function. NeoReviews. 2004;5:e252–e61.
-
- James JL, Srinivasan S, Alexander M, et al. Can we fix it? Evaluating the potential of placental stem cells for the treatment of pregnancy disorders. Placenta. 2014;35:77–84. - PubMed
-
- Huppertz B. Trophoblast differentiation, fetal growth restriction and preeclampsia. Pregnancy Hypertens. 2011;1:79–86. - PubMed
-
- Vlahovic M, Bulic-Jakus F, Juric-Lekic G, et al. Changes in the placenta and in the rat embryo caused by the demethylating agent 5-azacytidine. Int J Dev Biol. 1999;43:843–846. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
