The RNA helicase DDX3X is an essential mediator of innate antimicrobial immunity
- PMID: 30475900
- PMCID: PMC6283616
- DOI: 10.1371/journal.ppat.1007397
The RNA helicase DDX3X is an essential mediator of innate antimicrobial immunity
Abstract
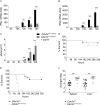
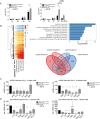
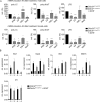
DExD/H box RNA helicases, such as the RIG-I-like receptors (RLR), are important components of the innate immune system. Here we demonstrate a pivotal and sex-specific role for the heterosomal isoforms of the DEAD box RNA helicase DDX3 in the immune system. Mice lacking DDX3X during hematopoiesis showed an altered leukocyte composition in bone marrow and spleen and a striking inability to combat infection with Listeria monocytogenes. Alterations in innate immune responses resulted from decreased effector cell availability and function as well as a sex-dependent impairment of cytokine synthesis. Thus, our data provide further in vivo evidence for an essential contribution of a non-RLR DExD/H RNA helicase to innate immunity and suggest it may contribute to sex-related differences in resistance to microbes and resilience to inflammatory disease.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






References
-
- Goubau D, Deddouche S, Reis E Sousa C. Cytosolic sensing of viruses. Immunity. 2013;38: 855–869. 10.1016/j.immuni.2013.05.007 - DOI - PMC - PubMed
-
- Paludan SR, Bowie AG. Immune sensing of DNA. Immunity. 2013;38: 870–880. 10.1016/j.immuni.2013.05.004 - DOI - PMC - PubMed
-
- Stavrou S, Aguilera AN, Blouch K, Ross SR. DDX41 Recognizes RNA/DNA Retroviral Reverse Transcripts and Is Critical for In Vivo Control of Murine Leukemia Virus Infection. MBio. American Society for Microbiology; 2018;9: e00923–18. 10.1128/mBio.00923-18 - DOI - PMC - PubMed
-
- Soulat D, Burckstummer T, Westermayer S, Goncalves A, Bauch A, Stefanovic A, et al. The DEAD-box helicase DDX3X is a critical component of the TANK-binding kinase 1-dependent innate immune response. EMBO J. 2008; 27: 2135–2146. 10.1038/emboj.2008.126 - DOI - PMC - PubMed
-
- Schröder M, Baran M, Bowie AG. Viral targeting of DEAD box protein 3 reveals its role in TBK1/IKKepsilon-mediated IRF activation. EMBO J. 2008; 27: 2147–2157. 10.1038/emboj.2008.143 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

