The genome of an underwater architect, the caddisfly Stenopsyche tienmushanensis Hwang (Insecta: Trichoptera)
- PMID: 30476205
- PMCID: PMC6302954
- DOI: 10.1093/gigascience/giy143
The genome of an underwater architect, the caddisfly Stenopsyche tienmushanensis Hwang (Insecta: Trichoptera)
Abstract
Background: Caddisflies (Insecta: Trichoptera) are a highly adapted freshwater group of insects split from a common ancestor with Lepidoptera. They are the most diverse (>16,000 species) of the strictly aquatic insect orders and are widely employed as bio-indicators in water quality assessment and monitoring. Among the numerous adaptations to aquatic habitats, caddisfly larvae use silk and materials from the environment (e.g., stones, sticks, leaf matter) to build composite structures such as fixed retreats and portable cases. Understanding how caddisflies have adapted to aquatic habitats will help explain the evolution and subsequent diversification of the group.
Findings: We sequenced a retreat-builder caddisfly Stenopsyche tienmushanensis Hwang and assembled a high-quality genome from both Illumina and Pacific Biosciences (PacBio) sequencing. In total, 601.2 M Illumina reads (90.2 Gb) and 16.9 M PacBio subreads (89.0 Gb) were generated. The 451.5 Mb assembled genome has a contig N50 of 1.29 M, has a longest contig of 4.76 Mb, and covers 97.65% of the 1,658 insect single-copy genes as assessed by Benchmarking Universal Single-Copy Orthologs. The genome comprises 36.76% repetitive elements. A total of 14,672 predicted protein-coding genes were identified. The genome revealed gene expansions in specific groups of the cytochrome P450 family and olfactory binding proteins, suggesting potential genomic features associated with pollutant tolerance and mate finding. In addition, the complete gene complex of the highly repetitive H-fibroin, the major protein component of caddisfly larval silk, was assembled.
Conclusions: We report the draft genome of Stenopsyche tienmushanensis, the highest-quality caddisfly genome so far. The genome information will be an important resource for the study of caddisflies and may shed light on the evolution of aquatic insects.
Figures

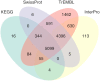
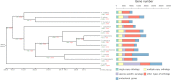



References
-
- Morse JC. The Trichoptera world checklist. Zoosymposia. 2011;5(1):372–80.
-
- Misof B, Liu S, Meusemann K et al. . Phylogenomics resolves the timing and pattern of insect evolution. Science. 2014;346(6210):763–7. - PubMed
-
- Resh VH, Unzicker JD. Water quality monitoring and aquatic organisms: the importance of species identification. J Water Pollut Control Fed. 1975;47(1):9–19. - PubMed
-
- Holzenthal R, Blahnik R, Kjer K et al. . An update on the phylogeny of caddisflies (Trichoptera). In: Proceedings of the 12th International Symposium on Trichoptera, The Caddis Press, Columbus, Ohio, 2007, pp. 143–53.
-
- Holzenthal RW, Thomson RE, Ríos-Touma B. Order Trichoptera. Thorp and Covich's Freshwater Invertebrates, Fourth Edition Elsevier; 2015. p. 965–1002.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

