The BioGRID interaction database: 2019 update
- PMID: 30476227
- PMCID: PMC6324058
- DOI: 10.1093/nar/gky1079
The BioGRID interaction database: 2019 update
Abstract
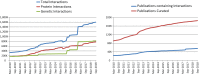
The Biological General Repository for Interaction Datasets (BioGRID: https://thebiogrid.org) is an open access database dedicated to the curation and archival storage of protein, genetic and chemical interactions for all major model organism species and humans. As of September 2018 (build 3.4.164), BioGRID contains records for 1 598 688 biological interactions manually annotated from 55 809 publications for 71 species, as classified by an updated set of controlled vocabularies for experimental detection methods. BioGRID also houses records for >700 000 post-translational modification sites. BioGRID now captures chemical interaction data, including chemical-protein interactions for human drug targets drawn from the DrugBank database and manually curated bioactive compounds reported in the literature. A new dedicated aspect of BioGRID annotates genome-wide CRISPR/Cas9-based screens that report gene-phenotype and gene-gene relationships. An extension of the BioGRID resource called the Open Repository for CRISPR Screens (ORCS) database (https://orcs.thebiogrid.org) currently contains over 500 genome-wide screens carried out in human or mouse cell lines. All data in BioGRID is made freely available without restriction, is directly downloadable in standard formats and can be readily incorporated into existing applications via our web service platforms. BioGRID data are also freely distributed through partner model organism databases and meta-databases.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

