piRNA-Guided Genome Defense: From Biogenesis to Silencing
- PMID: 30476449
- PMCID: PMC10784713
- DOI: 10.1146/annurev-genet-120417-031441
piRNA-Guided Genome Defense: From Biogenesis to Silencing
Abstract
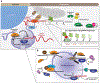
PIWI-interacting RNAs (piRNAs) and their associated PIWI clade Argonaute proteins constitute the core of the piRNA pathway. In gonadal cells, this conserved pathway is crucial for genome defense, and its main function is to silence transposable elements. This is achieved through posttranscriptional and transcriptional gene silencing. Precursors that give rise to piRNAs require specialized transcription and transport machineries because piRNA biogenesis is a cytoplasmic process. The ping-pong cycle, a posttranscriptional silencing mechanism, combines the cleavage-dependent silencing of transposon RNAs with piRNA production. PIWI proteins also function in the nucleus, where they scan for nascent target transcripts with sequence complementarity, instructing transcriptional silencing and deposition of repressive chromatin marks at transposon loci. Although studies have revealed numerous factors that participate in each branch of the piRNA pathway, the precise molecular roles of these factors often remain unclear. In this review, we summarize our current understanding of the mechanisms involved in piRNA biogenesis and function.
Keywords: PIWI proteins; piRNA biogenesis; piRNA clusters; ping-pong loop; posttranscriptional gene silencing; transcriptional gene silencing; transposon control.
Figures


References
-
- Akkouche A, Mugat B, Barckmann B, Varela-Chavez C, Li B, et al. 2017. Piwi is required during Drosophila embryogenesis to license dual-strand piRNA clusters for transposon repression in adult ovaries. Mol. Cell 66:411–19.e4 - PubMed
-
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, et al. 2006. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442:203–207 - PubMed
-
- Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, et al. 2003. The small RNA profile during Drosophila melanogaster development. Dev. Cell 5:337–50 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

