The microbiome in patients with atopic dermatitis
- PMID: 30476499
- PMCID: PMC7163929
- DOI: 10.1016/j.jaci.2018.11.015
The microbiome in patients with atopic dermatitis
Erratum in
-
Corrigendum.J Allergy Clin Immunol. 2019 Apr;143(4):1660. doi: 10.1016/j.jaci.2019.01.022. J Allergy Clin Immunol. 2019. PMID: 30954089 No abstract available.
Abstract
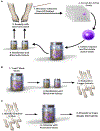
As an interface with the environment, the skin is a complex ecosystem colonized by many microorganisms that coexist in an established balance. The cutaneous microbiome inhibits colonization with pathogens, such as Staphylococcus aureus, and is a crucial component for function of the epidermal barrier. Moreover, crosstalk between commensals and the immune system is now recognized because microorganisms can modulate both innate and adaptive immune responses. Host-commensal interactions also have an effect on the developing immune system in infants and, subsequently, the occurrence of diseases, such as asthma and atopic dermatitis (AD). Later in life, the cutaneous microbiome contributes to the development and course of skin disease. Accordingly, in patients with AD, a decrease in microbiome diversity correlates with disease severity and increased colonization with pathogenic bacteria, such as S aureus. Early clinical studies suggest that topical application of commensal organisms (eg, Staphylococcus hominis or Roseomonas mucosa) reduces AD severity, which supports an important role for commensals in decreasing S aureus colonization in patients with AD. Advancing knowledge of the cutaneous microbiome and its function in modulating the course of skin disorders, such as AD, might result in novel therapeutic strategies.
Keywords: Atopic dermatitis; Staphylococcus aureus; biotherapy; commensal; host-microbiome interaction; immune regulation; microbiome.
Copyright © 2018 The Authors. Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Byrd AL, Belkaid Y, Segre JA. The human skin microbiome. Nat Rev Microbiol 2018;16:143–55. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

