UrdA Controls Secondary Metabolite Production and the Balance between Asexual and Sexual Development in Aspergillus nidulans
- PMID: 30477161
- PMCID: PMC6316066
- DOI: 10.3390/genes9120570
UrdA Controls Secondary Metabolite Production and the Balance between Asexual and Sexual Development in Aspergillus nidulans
Abstract
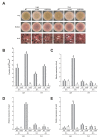
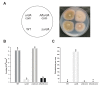
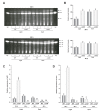
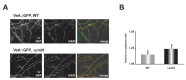
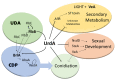
The genus Aspergillus includes important plant pathogens, opportunistic human pathogens and mycotoxigenic fungi. In these organisms, secondary metabolism and morphogenesis are subject to a complex genetic regulation. Here we functionally characterized urdA, a gene encoding a putative helix-loop-helix (HLH)-type regulator in the model fungus Aspergillus nidulans. urdA governs asexual and sexual development in strains with a wild-type veA background; absence of urdA resulted in severe morphological alterations, with a significant reduction of conidial production and an increase in cleistothecial formation, even in the presence of light, a repressor of sex. The positive effect of urdA on conidiation is mediated by the central developmental pathway (CDP). However, brlA overexpression was not sufficient to restore wild-type conidiation in the ΔurdA strain. Heterologous complementation of ΔurdA with the putative Aspergillus flavus urdA homolog also failed to rescue conidiation wild-type levels, indicating that both genes perform different functions, probably reflected by key sequence divergence. UrdA also represses sterigmatocystin (ST) toxin production in the presence of light by affecting the expression of aflR, the activator of the ST gene cluster. Furthermore, UrdA regulates the production of several unknown secondary metabolites, revealing a broader regulatory scope. Interestingly, UrdA affects the abundance and distribution of the VeA protein in hyphae, and our genetics studies indicated that veA appears epistatic to urdA regarding ST production. However, the distinct fluffy phenotype of the ΔurdAΔveA double mutant suggests that both regulators conduct independent developmental roles. Overall, these results suggest that UrdA plays a pivotal role in the coordination of development and secondary metabolism in A. nidulans.
Keywords: Aspergillus nidulans; UrdA; brlA; central developmental pathway; epistasis; morphological development; mycotoxin; secondary metabolism; sterigmatocystin; transcription factor; veA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
LinkOut - more resources
Full Text Sources

