Genomic Islands Confer Heavy Metal Resistance in Mucilaginibacter kameinonensis and Mucilaginibacter rubeus Isolated from a Gold/Copper Mine
- PMID: 30477188
- PMCID: PMC6316836
- DOI: 10.3390/genes9120573
Genomic Islands Confer Heavy Metal Resistance in Mucilaginibacter kameinonensis and Mucilaginibacter rubeus Isolated from a Gold/Copper Mine
Abstract
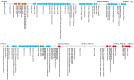
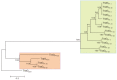
Heavy metals (HMs) are compounds that can be hazardous and impair growth of living organisms. Bacteria have evolved the capability not only to cope with heavy metals but also to detoxify polluted environments. Three heavy metal-resistant strains of Mucilaginibacer rubeus and one of Mucilaginibacter kameinonensis were isolated from the gold/copper Zijin mining site, Longyan, Fujian, China. These strains were shown to exhibit high resistance to heavy metals with minimal inhibitory concentration reaching up to 3.5 mM Cu(II), 21 mM Zn(II), 1.2 mM Cd(II), and 10.0 mM As(III). Genomes of the four strains were sequenced by Illumina. Sequence analyses revealed the presence of a high abundance of heavy metal resistance (HMR) determinants. One of the strain, M. rubeus P2, carried genes encoding 6 putative PIB-1-ATPase, 5 putative PIB-3-ATPase, 4 putative Zn(II)/Cd(II) PIB-4 type ATPase, and 16 putative resistance-nodulation-division (RND)-type metal transporter systems. Moreover, the four genomes contained a high abundance of genes coding for putative metal binding chaperones. Analysis of the close vicinity of these HMR determinants uncovered the presence of clusters of genes potentially associated with mobile genetic elements. These loci included genes coding for tyrosine recombinases (integrases) and subunits of mating pore (type 4 secretion system), respectively allowing integration/excision and conjugative transfer of numerous genomic islands. Further in silico analyses revealed that their genetic organization and gene products resemble the Bacteroides integrative and conjugative element CTnDOT. These results highlight the pivotal role of genomic islands in the acquisition and dissemination of adaptive traits, allowing for rapid adaption of bacteria and colonization of hostile environments.
Keywords: CTnDOT; Mucilaginibacer rubeus; Mucilaginibacter kameinonensis; draft genome sequence; evolution; genomic island; heavy metal resistance.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Extrapolymeric substances (EPS) in Mucilaginibacter rubeus P2 displayed efficient metal(loid) bio-adsorption and production was induced by copper and zinc.Chemosphere. 2022 Mar;291(Pt 1):132712. doi: 10.1016/j.chemosphere.2021.132712. Epub 2021 Oct 26. Chemosphere. 2022. PMID: 34715104
-
New mobile genetic elements in Cupriavidus metallidurans CH34, their possible roles and occurrence in other bacteria.Antonie Van Leeuwenhoek. 2009 Aug;96(2):205-26. doi: 10.1007/s10482-009-9345-4. Epub 2009 Apr 24. Antonie Van Leeuwenhoek. 2009. PMID: 19390985
-
Comparative Insights Into the Complete Genome Sequence of Highly Metal Resistant Cupriavidus metallidurans Strain BS1 Isolated From a Gold-Copper Mine.Front Microbiol. 2020 Feb 7;11:47. doi: 10.3389/fmicb.2020.00047. eCollection 2020. Front Microbiol. 2020. PMID: 32117100 Free PMC article.
-
Cupriavidus metallidurans: evolution of a metal-resistant bacterium.Antonie Van Leeuwenhoek. 2009 Aug;96(2):115-39. doi: 10.1007/s10482-008-9284-5. Epub 2008 Oct 1. Antonie Van Leeuwenhoek. 2009. PMID: 18830684 Review.
-
From industrial sites to environmental applications with Cupriavidus metallidurans.Antonie Van Leeuwenhoek. 2009 Aug;96(2):247-58. doi: 10.1007/s10482-009-9361-4. Epub 2009 Jul 7. Antonie Van Leeuwenhoek. 2009. PMID: 19582590 Review.
Cited by
-
Complete genome sequence of Mucilaginibacter gossypii P3, a heavy metal(loid)-resistant bacterium, isolated from a gold and copper mine.Microbiol Resour Announc. 2023 Nov 16;12(11):e0017223. doi: 10.1128/MRA.00172-23. Epub 2023 Oct 10. Microbiol Resour Announc. 2023. PMID: 37815359 Free PMC article.
-
Recent advances in exploring the heavy metal(loid) resistant microbiome.Comput Struct Biotechnol J. 2020 Dec 14;19:94-109. doi: 10.1016/j.csbj.2020.12.006. eCollection 2021. Comput Struct Biotechnol J. 2020. PMID: 33425244 Free PMC article. Review.
-
Isolation and pan-genome analysis of Acinetobacter junii SC22, a heavy metal(loid)s resistant and plant growth promoting bacterium, from the Zijin Gold and Copper mine.Biometals. 2025 Jul 21. doi: 10.1007/s10534-025-00724-3. Online ahead of print. Biometals. 2025. PMID: 40690081
-
Azolla filiculoides L. as a source of metal-tolerant microorganisms.PLoS One. 2020 May 6;15(5):e0232699. doi: 10.1371/journal.pone.0232699. eCollection 2020. PLoS One. 2020. PMID: 32374760 Free PMC article.
-
Adaptation to metal(loid)s in strain Mucilaginibacter rubeus P2 involves novel arsenic resistance genes and mechanisms.J Hazard Mater. 2024 Jan 15;462:132796. doi: 10.1016/j.jhazmat.2023.132796. Epub 2023 Oct 16. J Hazard Mater. 2024. PMID: 37865075 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

