Extraction of Information Related to Adverse Drug Events from Electronic Health Record Notes: Design of an End-to-End Model Based on Deep Learning
- PMID: 30478023
- PMCID: PMC6288593
- DOI: 10.2196/12159
Extraction of Information Related to Adverse Drug Events from Electronic Health Record Notes: Design of an End-to-End Model Based on Deep Learning
Abstract
Background: Pharmacovigilance and drug-safety surveillance are crucial for monitoring adverse drug events (ADEs), but the main ADE-reporting systems such as Food and Drug Administration Adverse Event Reporting System face challenges such as underreporting. Therefore, as complementary surveillance, data on ADEs are extracted from electronic health record (EHR) notes via natural language processing (NLP). As NLP develops, many up-to-date machine-learning techniques are introduced in this field, such as deep learning and multi-task learning (MTL). However, only a few studies have focused on employing such techniques to extract ADEs.
Objective: We aimed to design a deep learning model for extracting ADEs and related information such as medications and indications. Since extraction of ADE-related information includes two steps-named entity recognition and relation extraction-our second objective was to improve the deep learning model using multi-task learning between the two steps.
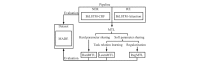
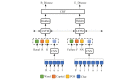
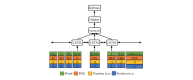
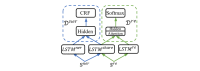
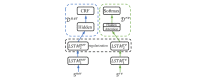
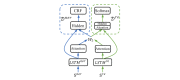
Methods: We employed the dataset from the Medication, Indication and Adverse Drug Events (MADE) 1.0 challenge to train and test our models. This dataset consists of 1089 EHR notes of cancer patients and includes 9 entity types such as Medication, Indication, and ADE and 7 types of relations between these entities. To extract information from the dataset, we proposed a deep-learning model that uses a bidirectional long short-term memory (BiLSTM) conditional random field network to recognize entities and a BiLSTM-Attention network to extract relations. To further improve the deep-learning model, we employed three typical MTL methods, namely, hard parameter sharing, parameter regularization, and task relation learning, to build three MTL models, called HardMTL, RegMTL, and LearnMTL, respectively.
Results: Since extraction of ADE-related information is a two-step task, the result of the second step (ie, relation extraction) was used to compare all models. We used microaveraged precision, recall, and F1 as evaluation metrics. Our deep learning model achieved state-of-the-art results (F1=65.9%), which is significantly higher than that (F1=61.7%) of the best system in the MADE1.0 challenge. HardMTL further improved the F1 by 0.8%, boosting the F1 to 66.7%, whereas RegMTL and LearnMTL failed to boost the performance.
Conclusions: Deep learning models can significantly improve the performance of ADE-related information extraction. MTL may be effective for named entity recognition and relation extraction, but it depends on the methods, data, and other factors. Our results can facilitate research on ADE detection, NLP, and machine learning.
Keywords: adverse drug event; deep learning; multi-task learning; named entity recognition; natural language processing; relation extraction.
©Fei Li, Weisong Liu, Hong Yu. Originally published in JMIR Medical Informatics (http://medinform.jmir.org), 26.11.2018.
Conflict of interest statement
Conflicts of Interest: None declared.
Figures
References
-
- Bates DW, Cullen DJ, Laird N, Petersen LA, Small SD, Servi D, Laffel G, Sweitzer BJ, Shea BF, Hallisey R. Incidence of adverse drug events and potential adverse drug events. Implications for prevention. ADE Prevention Study Group. JAMA. 1995 Jul 05;274(1):29–34. - PubMed
-
- Fattinger K, Roos M, Vergères P, Holenstein C, Kind B, Masche U, Stocker DN, Braunschweig S, Kullak-Ublick GA, Galeazzi RL, Follath F, Gasser T, Meier PJ. Epidemiology of drug exposure and adverse drug reactions in two swiss departments of internal medicine. Br J Clin Pharmacol. 2000 Feb;49(2):158–67. https://onlinelibrary.wiley.com/resolve/openurl?genre=article&sid=nlm:pu... bcp132 - PMC - PubMed
-
- Classen DC, Pestotnik SL, Evans RS, Lloyd JF, Burke JP. Adverse drug events in hospitalized patients. Excess length of stay, extra costs, and attributable mortality. JAMA. 1997;277(4):301–6. - PubMed
-
- Bates DW, Spell N, Cullen DJ, Burdick E, Laird N, Petersen LA, Small SD, Sweitzer BJ, Leape LL. The costs of adverse drug events in hospitalized patients. Adverse Drug Events Prevention Study Group. JAMA. 1997;277(4):307–11. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

