Structures of AMP-activated protein kinase bound to novel pharmacological activators in phosphorylated, non-phosphorylated, and nucleotide-free states
- PMID: 30478170
- PMCID: PMC6341387
- DOI: 10.1074/jbc.RA118.004883
Structures of AMP-activated protein kinase bound to novel pharmacological activators in phosphorylated, non-phosphorylated, and nucleotide-free states
Abstract
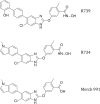
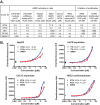
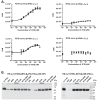
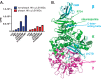
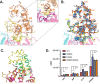
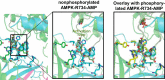
AMP-activated protein kinase (AMPK) is an attractive therapeutic target for managing metabolic diseases. A class of pharmacological activators, including Merck 991, binds the AMPK ADaM site, which forms the interaction surface between the kinase domain (KD) of the α-subunit and the carbohydrate-binding module (CBM) of the β-subunit. Here, we report the development of two new 991-derivative compounds, R734 and R739, which potently activate AMPK in a variety of cell types, including β2-specific skeletal muscle cells. Surprisingly, we found that they have only minor effects on direct kinase activity of the recombinant α1β2γ1 isoform yet robustly enhance protection against activation loop dephosphorylation. This mode of activation is reminiscent of that of ADP, which activates AMPK by binding to the nucleotide-binding sites in the γ-subunit, more than 60 Å away from the ADaM site. To understand the mechanisms of full and partial AMPK activation, we determined the crystal structures of fully active phosphorylated AMPK α1β1γ1 bound to AMP and R734/R739 as well as partially active nonphosphorylated AMPK bound to R734 and AMP and phosphorylated AMPK bound to R734 in the absence of added nucleotides at <3-Å resolution. These structures and associated analyses identified a novel conformational state of the AMPK autoinhibitory domain associated with partial kinase activity and provide new insights into phosphorylation-dependent activation loop stabilization in AMPK.
Keywords: ADaM site; AMP; AMP-activated kinase (AMPK); CBS3; R734; R739; X-ray crystallography; activation loop phosphorylation; energy sensor; hydrogen exchange mass spectrometry; metabolic disorder; phosphorylation.
© 2019 Yan et al.
Conflict of interest statement
S. J. S., Y. L., and Y. H. are or have been employees and shareholders of Rigel Pharmaceuticals, Inc
Figures




References
-
- Stapleton D., Gao G., Michell B. J., Widmer J., Mitchelhill K., Teh T., House C. M., Witters L. A., and Kemp B. E. (1994) Mammalian 5′-AMP-activated protein kinase non-catalytic subunits are homologs of proteins that interact with yeast Snf1 protein kinase. J. Biol. Chem. 269, 29343–29346 - PubMed
-
- Cokorinos E. C., Delmore J., Reyes A. R., Albuquerque B., Kjøbsted R., Jørgensen N. O., Tran J. L., Jatkar A., Cialdea K., Esquejo R. M., Meissen J., Calabrese M. F., Cordes J., Moccia R., Tess D., et al. (2017) Activation of skeletal muscle AMPK promotes glucose disposal and glucose lowering in non-human primates and mice. Cell Metab. 25, 1147–1159.e10 10.1074/jbc.RA117.001068 - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

