CBX2 identified as driver of anoikis escape and dissemination in high grade serous ovarian cancer
- PMID: 30478317
- PMCID: PMC6255906
- DOI: 10.1038/s41389-018-0103-1
CBX2 identified as driver of anoikis escape and dissemination in high grade serous ovarian cancer
Abstract
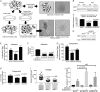
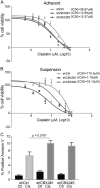
High grade serous ovarian carcinoma (HGSOC) is often diagnosed at an advanced stage. Chromobox 2 (CBX2), a polycomb repressor complex subunit, plays an oncogenic role in other cancers, but little is known about its role in HGSOC. We hypothesize that CBX2 upregulation promotes HGSOC via induction of a stem-like transcriptional profile and inhibition of anoikis. Examination of Gene Expression Omnibus (GEO) datasets and The Cancer Genome Atlas (TCGA) established that increased CBX2 expression conveyed chemoresistance and worse disease-free and overall survival. In primary HGSOC tumors, we observed CBX2 expression was significantly elevated compared to benign counterparts. In HGSOC cell lines, forced suspension promoted CBX2 expression. Subsequently, CBX2 knockdown inhibited anchorage-independent proliferation and potentiated anoikis-dependent apoptosis. Furthermore, CBX2 knockdown re-sensitized cells to platinum-based chemotherapy. Forced suspension promoted increased ALDH activity and ALDH3A1 expression and CBX2 knockdown led to a decrease in both ALDH activity and ALDH3A1 expression. Investigation of CBX2 expression on a HGSOC tissue microarray revealed CBX2 expression was apparent in both primary and metastatic tissues. CBX2 is an important regulator of stem-ness, anoikis escape, HGSOC dissemination, and chemoresistance and potentially serves as a novel therapeutic target.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

