The interferon-inducible isoform of NCOA7 inhibits endosome-mediated viral entry
- PMID: 30478388
- PMCID: PMC6329445
- DOI: 10.1038/s41564-018-0273-9
The interferon-inducible isoform of NCOA7 inhibits endosome-mediated viral entry
Erratum in
-
Author Correction: The interferon-inducible isoform of NCOA7 inhibits endosome-mediated viral entry.Nat Microbiol. 2019 Mar;4(3):539. doi: 10.1038/s41564-019-0366-0. Nat Microbiol. 2019. PMID: 30670794
Abstract
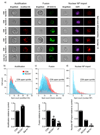
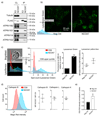
Interferons (IFNs) mediate cellular defence against viral pathogens by upregulation of IFN-stimulated genes whose products interact with viral components or alter cellular physiology to suppress viral replication1-3. Among the IFN-stimulated genes that can inhibit influenza A virus (IAV)4 are the myxovirus resistance 1 GTPase5 and IFN-induced transmembrane protein 3 (refs 6,7). Here, we use ectopic expression and gene knockout to demonstrate that the IFN-inducible 219-amino acid short isoform of human nuclear receptor coactivator 7 (NCOA7) is an inhibitor of IAV as well as other viruses that enter the cell by endocytosis, including hepatitis C virus. NCOA7 interacts with the vacuolar H+-ATPase (V-ATPase) and its expression promotes cytoplasmic vesicle acidification, lysosomal protease activity and the degradation of endocytosed antigen. Step-wise dissection of the IAV entry pathway demonstrates that NCOA7 inhibits fusion of the viral and endosomal membranes and subsequent nuclear translocation of viral ribonucleoproteins. Therefore, NCOA7 provides a mechanism for immune regulation of endolysosomal physiology that not only suppresses viral entry into the cytosol from this compartment but may also regulate other V-ATPase-associated cellular processes, such as physiological adjustments to nutritional status, or the maturation and function of antigen-presenting cells.
Conflict of interest statement
The authors have no conflicts of interest to declare in relation to this manuscript.
Figures

References
-
- Randall RE, Goodbourn S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. The Journal of general virology. 2008;89(Pt 1):1–47. - PubMed
-
- Haller O, Kochs G. Human MxA protein: an interferon-induced dynamin-like GTPase with broad antiviral activity. Journal of interferon & cytokine research : the official journal of the International Society for Interferon and Cytokine Research. 2011;31(1):79–87. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

