Vasculogenic mimicry formation in EBV-associated epithelial malignancies
- PMID: 30479336
- PMCID: PMC6258759
- DOI: 10.1038/s41467-018-07308-5
Vasculogenic mimicry formation in EBV-associated epithelial malignancies
Retraction in
-
Retraction Note: Vasculogenic mimicry formation in EBV-associated epithelial malignancies.Nat Commun. 2025 Oct 27;16(1):9440. doi: 10.1038/s41467-025-65413-8. Nat Commun. 2025. PMID: 41145499 Free PMC article. No abstract available.
Abstract
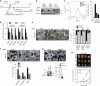
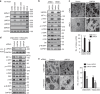
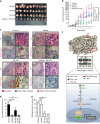
Epstein-Barr virus (EBV)-associated epithelial cancers, including nasopharyngeal carcinoma (NPC) and approximately 10% of gastric cancers, termed EBVaGC, represent 80% of all EBV-related malignancies. However, the exact role of EBV in epithelial cancers remains elusive. Here, we report that EBV functions in vasculogenic mimicry (VM). Epithelial cancer cells infected with EBV develop tumor vascular networks that correlate with tumor growth, which is different from endothelial-derived angiogenic vessels and is VEGF-independent. Mechanistically, activation of the PI3K/AKT/mTOR/HIF-1α signaling cascade, which is partly mediated by LMP2A, is responsible for EBV-induced VM formation. Both xenografts and clinical samples of NPC and EBVaGC exhibit VM histologically, which are correlated with AKT and HIF-1α activation. Furthermore, although anti-VEGF monotherapy shows limited effects, potent synergistic antitumor activities are achieved by combination therapy with VEGF and HIF-1α-targeted agents. Our findings suggest that EBV creates plasticity in epithelial cells to express endothelial phenotype and provides a novel EBV-targeted antitumor strategy.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Young, L. S., Yap, L. F. & Murray, P. G. Epstein-Barr virus: more than 50 years old and still providing surprises. Nat. Rev. Cancer16, 789–802 (2016). - PubMed
-
- Chang, E. T. & Adami, H. O. The enigmatic epidemiology of nasopharyngeal carcinoma. Cancer Epidemiol. Biomark. Prev.15, 1765–1777 (2006). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

