Modeling craniofacial development reveals spatiotemporal constraints on robust patterning of the mandibular arch
- PMID: 30481168
- PMCID: PMC6258504
- DOI: 10.1371/journal.pcbi.1006569
Modeling craniofacial development reveals spatiotemporal constraints on robust patterning of the mandibular arch
Abstract
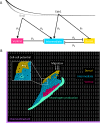
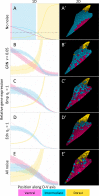
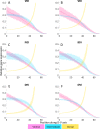
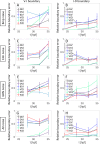
How does pattern formation occur accurately when confronted with tissue growth and stochastic fluctuations (noise) in gene expression? Dorso-ventral (D-V) patterning of the mandibular arch specifies upper versus lower jaw skeletal elements through a combination of Bone morphogenetic protein (Bmp), Endothelin-1 (Edn1), and Notch signaling, and this system is highly robust. We combine NanoString experiments of early D-V gene expression with live imaging of arch development in zebrafish to construct a computational model of the D-V mandibular patterning network. The model recapitulates published genetic perturbations in arch development. Patterning is most sensitive to changes in Bmp signaling, and the temporal order of gene expression modulates the response of the patterning network to noise. Thus, our integrated systems biology approach reveals non-intuitive features of the complex signaling system crucial for craniofacial development, including novel insights into roles of gene expression timing and stochasticity in signaling and gene regulation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Lawrence PA, Crick FHC, Munro M. A Gradient of Positional Information in an Insect, Rhodnius. J Cell Sci. 1972. November 1;11(3):815–53. - PubMed
-
- Thompson DW. On growth and form Cambridge [Eng.]: University press; 1917. xv, 793.
-
- Turing AM. The Chemical Basis of Morphogenesis. Philos Trans R Soc Lond B Biol Sci. 1952. August 14;237(641):37–72.
-
- Wolpert L. Positional information and the spatial pattern of cellular differentiation. J Theor Biol. 1969. October 1;25(1):1–47. - PubMed
-
- Alon U. Network motifs: theory and experimental approaches. Nat Rev Genet. 2007. June;8(6):450–61. 10.1038/nrg2102 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

