Dual positive and negative control of Chlamydomonas PII signal transduction protein expression by nitrate/nitrite and NO via the components of nitric oxide cycle
- PMID: 30482162
- PMCID: PMC6258461
- DOI: 10.1186/s12870-018-1540-x
Dual positive and negative control of Chlamydomonas PII signal transduction protein expression by nitrate/nitrite and NO via the components of nitric oxide cycle
Abstract
Background: The PII proteins constitute a large superfamily, present in all domains of life. Until now, PII proteins research in Chloroplastida (green algae and land plants) has mainly focused on post-translation regulation of these signal transductors. Emerging evidence suggests that PII level is tightly controlled with regard to the nitrogen source and the physiological state of cells.
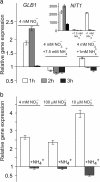
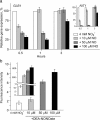
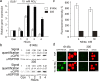
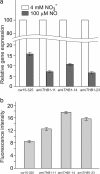
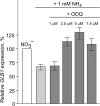
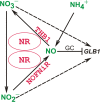
Result: Here we identify that a balance of positive (nitrate and nitrite) and negative (nitric oxide) signals regulates Chlamydomonas GLB1. We found that PII expression is downregulated by ammonium through a nitric oxide (NO)-dependent mechanism. We show that nitrate reductase (NR) and its partner, truncated hemoglobin 1 (THB1), participate in a signaling pathway for dual control of GLB1 expression. Moreover, NO dependent guanilate cyclase appeared to be involved in the negative control of GLB1 transcription.
Conclusion: This study has revealed the existence of the complex GLB1 control at transcription level, which is dependent on nitrogen source. Importantly, we found that GLB1 gene expression pattern is very similar to that observed for nitrate assimilation genes, suggesting interconnecting/coordinating PII-dependent and nitrate assimilation pathways.
Keywords: Chlamydomonas reinhardtii; NO signaling; Nitrate; Nitrite; PII signal transduction protein; Truncated hemoglobin.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

Similar articles
-
THB1 regulates nitrate reductase activity and THB1 and THB2 transcription differentially respond to NO and the nitrate/ammonium balance in Chlamydomonas.Plant Signal Behav. 2015;10(8):e1042638. doi: 10.1080/15592324.2015.1042638. Plant Signal Behav. 2015. PMID: 26252500 Free PMC article.
-
How Chlamydomonas handles nitrate and the nitric oxide cycle.J Exp Bot. 2017 May 1;68(10):2593-2602. doi: 10.1093/jxb/erw507. J Exp Bot. 2017. PMID: 28201747 Review.
-
Nitric oxide controls nitrate and ammonium assimilation in Chlamydomonas reinhardtii.J Exp Bot. 2013 Aug;64(11):3373-83. doi: 10.1093/jxb/ert175. J Exp Bot. 2013. PMID: 23918969
-
THB1, a truncated hemoglobin, modulates nitric oxide levels and nitrate reductase activity.Plant J. 2015 Feb;81(3):467-79. doi: 10.1111/tpj.12744. Plant J. 2015. PMID: 25494936
-
Nitrate Reductase Regulates Plant Nitric Oxide Homeostasis.Trends Plant Sci. 2017 Feb;22(2):163-174. doi: 10.1016/j.tplants.2016.12.001. Epub 2017 Jan 5. Trends Plant Sci. 2017. PMID: 28065651 Review.
Cited by
-
Advanced pathway engineering for phototrophic putrescine production.Plant Biotechnol J. 2022 Oct;20(10):1968-1982. doi: 10.1111/pbi.13879. Epub 2022 Jul 22. Plant Biotechnol J. 2022. PMID: 35748533 Free PMC article.
-
Role of Nitrate Reductase in NO Production in Photosynthetic Eukaryotes.Plants (Basel). 2019 Mar 6;8(3):56. doi: 10.3390/plants8030056. Plants (Basel). 2019. PMID: 30845759 Free PMC article. Review.
-
Nitrous Oxide Emissions from Nitrite Are Highly Dependent on Nitrate Reductase in the Microalga Chlamydomonas reinhardtii.Int J Mol Sci. 2022 Aug 20;23(16):9412. doi: 10.3390/ijms23169412. Int J Mol Sci. 2022. PMID: 36012676 Free PMC article.
-
N-Acetyl-L-glutamate Kinase of Chlamydomonas reinhardtii: In Vivo Regulation by PII Protein and Beyond.Int J Mol Sci. 2023 Aug 17;24(16):12873. doi: 10.3390/ijms241612873. Int J Mol Sci. 2023. PMID: 37629055 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

