dbMPIKT: a database of kinetic and thermodynamic mutant protein interactions
- PMID: 30482172
- PMCID: PMC6260753
- DOI: 10.1186/s12859-018-2493-7
dbMPIKT: a database of kinetic and thermodynamic mutant protein interactions
Abstract
Background: Protein-protein interactions (PPIs) play important roles in biological functions. Studies of the effects of mutants on protein interactions can provide further understanding of PPIs. Currently, many databases collect experimental mutants to assess protein interactions, but most of these databases are old and have not been updated for several years.
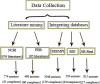
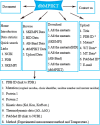
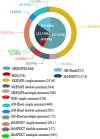
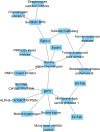
Results: To address this issue, we manually curated a kinetic and thermodynamic database of mutant protein interactions (dbMPIKT) that is freely accessible at our website. This database contains 5291 mutants in protein interactions collected from previous databases and the literature published within the last three years. Furthermore, some data analysis, such as mutation number, mutation type, protein pair source and network map construction, can be performed online.
Conclusion: Our work can promote the study on PPIs, and novel information can be mined from the new database. Our database is available in http://DeepLearner.ahu.edu.cn/web/dbMPIKT/ for use by all, including both academics and non-academics.
Keywords: Kinetic data; Mutants; Protein-protein interactions; Thermodynamic data.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
-
- Hu G, Xiao F, Li Y, Li Y, Vongsangnak W. Protein-protein Interface and disease: perspective from biomolecular networks. Adv Biochem Eng Biotechnol. 2017;160:57–74. - PubMed
-
- Zarei O, Hamzeh-Mivehroud M, Benvenuti S, Ustun-Alkan F, Dastmalchi S. Characterizing the hot spots involved in RON-MSP complex formation using in silico alanine scanning mutagenesis and molecular dynamics simulation. Advanced pharmaceutical bulletin. 2017;7:141–150. doi: 10.15171/apb.2017.018. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources