STING Promotes Homeostasis via Regulation of Cell Proliferation and Chromosomal Stability
- PMID: 30482772
- PMCID: PMC6445702
- DOI: 10.1158/0008-5472.CAN-18-1972
STING Promotes Homeostasis via Regulation of Cell Proliferation and Chromosomal Stability
Abstract
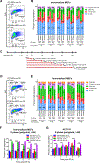
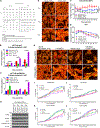
Given the integral role of stimulator of interferon genes (STING, TMEM173) in the innate immune response, its loss or impairment in cancer is thought to primarily affect antitumor immunity. Here we demonstrate a role for STING in the maintenance of cellular homeostasis through regulation of the cell cycle. Depletion of STING in human and murine cancer cells and tumors resulted in increased proliferation compared with wild-type controls. Microarray analysis revealed genes involved in cell-cycle regulation are differentially expressed in STINGko compared with WT MEFs. STING-mediated regulation of the cell cycle converged on NFκB- and p53-driven activation of p21. The absence of STING led to premature activation of cyclin-dependent kinase 1 (CDK1), early onset to S-phase and mitosis, and increased chromosome instability, which was enhanced by ionizing radiation. These results suggest a pivotal role for STING in maintaining cellular homeostasis and response to genotoxic stress. SIGNIFICANCE: These findings provide clear mechanistic understanding of the role of STING in cell-cycle regulation, which may be exploited in cancer therapy because most normal cells express STING, while many tumor cells do not.See related commentary by Gius and Zhu, p. 1295.
©2018 American Association for Cancer Research.
Conflict of interest statement
Figures



Comment in
-
Homeostatic Roles of STING in Cell Proliferation and Chromosomal Instability.Cancer Res. 2019 Apr 1;79(7):1295-1296. doi: 10.1158/0008-5472.CAN-19-0212. Cancer Res. 2019. PMID: 30936074
-
Expanding the Role of STING in Cellular Homeostasis and Transformation.Trends Cancer. 2019 Apr;5(4):195-197. doi: 10.1016/j.trecan.2019.02.001. Epub 2019 Feb 23. Trends Cancer. 2019. PMID: 30961826 Free PMC article.
References
-
- Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H, et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proceedings of the National Academy of Sciences of the United States of America 2009;106(49):20842–6 10.1073/pnas.0911267106. - DOI - PMC - PubMed
-
- Zhong B, Yang Y, Li S, Wang Y-Y, Li Y, Diao F, et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 2008;29(4):538–50. - PubMed
-
- Liu S, Cai X, Wu J, Cong Q, Chen X, Li T, et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science (New York, NY) 2015;347(6227):aaa2630. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

