Ribosomal Lesions Promote Oncogenic Mutagenesis
- PMID: 30482776
- PMCID: PMC7116100
- DOI: 10.1158/0008-5472.CAN-18-1987
Ribosomal Lesions Promote Oncogenic Mutagenesis
Abstract
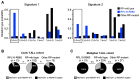
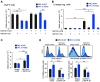
Ribosomopathies are congenital disorders caused by mutations in ribosomal proteins (RP) or assembly factors and are characterized by cellular hypoproliferation at an early stage. Paradoxically, many of these disorders have an elevated risk to progress to hyperproliferative cancer at a later stage. In addition, somatic RP mutations have recently been identified in various cancer types, for example, the recurrent RPL10-R98S mutation in T-cell acute lymphoblastic leukemia (T-ALL) and RPS15 mutations in chronic lymphocytic leukemia (CLL). We previously showed that RPL10-R98S promotes expression of oncogenes, but also induces a proliferative defect due to elevated oxidative stress. In this study, we demonstrate that this proliferation defect is eventually rescued by RPL10-R98S mouse lymphoid cells that acquire 5-fold more secondary mutations than RPL10-WT cells. The presence of RPL10-R98S and other RP mutations also correlated with a higher mutational load in patients with T-ALL, with an enrichment in NOTCH1-activating lesions. RPL10-R98S-associated cellular oxidative stress promoted DNA damage and impaired cell growth. Expression of NOTCH1 eliminated these phenotypes in RPL10-R98S cells, in part via downregulation of PKC-θ, with no effect on RPL10-WT cells. Patients with RP-mutant CLL also demonstrated a higher mutational burden, enriched for mutations that may diminish oxidative stress. We propose that oxidative stress due to ribosome dysfunction causes hypoproliferation and cellular insufficiency in ribosomopathies and RP-mutant cancer. This drives surviving cells, potentiated by genomic instability, to acquire rescuing mutations, which ultimately promote transition to hyperproliferation. SIGNIFICANCE: Ribosomal lesions cause oxidative stress and increase mutagenesis, promoting acquisition of rescuing mutations that stimulate proliferation.
©2018 American Association for Cancer Research.
Conflict of interest statement
Figures


References
-
- Dameshek W. Riddle: what do aplastic anemia, paroxysmal nocturnal hemoglobinuria (PNH) and “hypoplastic” leukemia have in common? Blood. 1967;30:251–254. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

