The Chaperone and Redox Properties of CnoX Chaperedoxins Are Tailored to the Proteostatic Needs of Bacterial Species
- PMID: 30482828
- PMCID: PMC6282202
- DOI: 10.1128/mBio.01541-18
The Chaperone and Redox Properties of CnoX Chaperedoxins Are Tailored to the Proteostatic Needs of Bacterial Species
Abstract
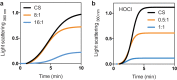
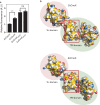
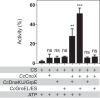
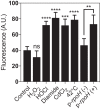
Hypochlorous acid (bleach), an oxidizing compound produced by neutrophils, turns the Escherichia coli chaperedoxin CnoX into a powerful holdase protecting its substrates from bleach-induced aggregation. CnoX is well conserved in bacteria, even in non-infectious species unlikely to encounter this oxidant, muddying the role of CnoX in these organisms. Here, we found that CnoX in the non-pathogenic aquatic bacterium Caulobacter crescentus functions as a holdase that efficiently protects 50 proteins from heat-induced aggregation. Remarkably, the chaperone activity of Caulobacter CnoX is constitutive. Like E. coli CnoX, Caulobacter CnoX transfers its substrates to DnaK/J/GrpE and GroEL/ES for refolding, indicating conservation of cooperation with GroEL/ES. Interestingly, Caulobacter CnoX exhibits thioredoxin oxidoreductase activity, by which it controls the redox state of 90 proteins. This function, which E. coli CnoX lacks, is likely welcome in a bacterium poorly equipped with antioxidant defenses. Thus, the redox and chaperone properties of CnoX chaperedoxins were fine-tuned during evolution to adapt these proteins to the specific needs of each species.IMPORTANCE How proteins are protected from stress-induced aggregation is a crucial question in biology and a long-standing mystery. While a long series of landmark studies have provided important contributions to our current understanding of the proteostasis network, key fundamental questions remain unsolved. In this study, we show that the intrinsic features of the chaperedoxin CnoX, a folding factor that combines chaperone and redox protective function, have been tailored during evolution to fit to the specific needs of their host. Whereas Escherichia coli CnoX needs to be activated by bleach, a powerful oxidant produced by our immune system, its counterpart in Caulobacter crescentus, a bacterium living in bleach-free environments, is a constitutive chaperone. In addition, the redox properties of E. coli and C. crescentus CnoX also differ to best contribute to their respective cellular redox homeostasis. This work demonstrates how proteins from the same family have evolved to meet the needs of their hosts.
Keywords: chaperones; oxidative stress; protein folding; thioredoxin.
Copyright © 2018 Goemans et al.
Figures


Similar articles
-
Stress-induced chaperones: a first line of defense against the powerful oxidant hypochlorous acid.F1000Res. 2019 Sep 23;8:F1000 Faculty Rev-1678. doi: 10.12688/f1000research.19517.1. eCollection 2019. F1000Res. 2019. PMID: 31583082 Free PMC article. Review.
-
CnoX Is a Chaperedoxin: A Holdase that Protects Its Substrates from Irreversible Oxidation.Mol Cell. 2018 May 17;70(4):614-627.e7. doi: 10.1016/j.molcel.2018.04.002. Epub 2018 May 10. Mol Cell. 2018. PMID: 29754824
-
Fort CnoX: Protecting Bacterial Proteins From Misfolding and Oxidative Damage.Front Mol Biosci. 2021 May 4;8:681932. doi: 10.3389/fmolb.2021.681932. eCollection 2021. Front Mol Biosci. 2021. PMID: 34017858 Free PMC article. Review.
-
A molecular device for the redox quality control of GroEL/ES substrates.Cell. 2023 Mar 2;186(5):1039-1049.e17. doi: 10.1016/j.cell.2023.01.013. Epub 2023 Feb 9. Cell. 2023. PMID: 36764293 Free PMC article.
-
The Thioredoxin Fold Protein (TFP2) from Extreme Acidophilic Leptospirillum sp. CF-1 Is a Chaperedoxin-like Protein That Prevents the Aggregation of Proteins under Oxidative Stress.Int J Mol Sci. 2024 Jun 24;25(13):6905. doi: 10.3390/ijms25136905. Int J Mol Sci. 2024. PMID: 39000017 Free PMC article.
Cited by
-
Surviving Reactive Chlorine Stress: Responses of Gram-Negative Bacteria to Hypochlorous Acid.Microorganisms. 2020 Aug 11;8(8):1220. doi: 10.3390/microorganisms8081220. Microorganisms. 2020. PMID: 32796669 Free PMC article. Review.
-
Stress-induced chaperones: a first line of defense against the powerful oxidant hypochlorous acid.F1000Res. 2019 Sep 23;8:F1000 Faculty Rev-1678. doi: 10.12688/f1000research.19517.1. eCollection 2019. F1000Res. 2019. PMID: 31583082 Free PMC article. Review.
-
Expression of RcrB confers resistance to hypochlorous acid in uropathogenic Escherichia coli.bioRxiv [Preprint]. 2023 Jun 1:2023.06.01.543251. doi: 10.1101/2023.06.01.543251. bioRxiv. 2023. Update in: J Bacteriol. 2023 Oct 26;205(10):e0006423. doi: 10.1128/jb.00064-23. PMID: 37398214 Free PMC article. Updated. Preprint.
-
The Chaperonin GroESL Facilitates Caulobacter crescentus Cell Division by Supporting the Functions of the Z-Ring Regulators FtsA and FzlA.mBio. 2021 May 4;12(3):e03564-20. doi: 10.1128/mBio.03564-20. mBio. 2021. PMID: 33947758 Free PMC article.
-
Redox-Mediated Inactivation of the Transcriptional Repressor RcrR is Responsible for Uropathogenic Escherichia coli's Increased Resistance to Reactive Chlorine Species.mBio. 2022 Oct 26;13(5):e0192622. doi: 10.1128/mbio.01926-22. Epub 2022 Sep 8. mBio. 2022. PMID: 36073817 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
