Supragingival Plaque Microbiome Ecology and Functional Potential in the Context of Health and Disease
- PMID: 30482830
- PMCID: PMC6282201
- DOI: 10.1128/mBio.01631-18
Supragingival Plaque Microbiome Ecology and Functional Potential in the Context of Health and Disease
Abstract
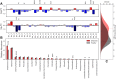
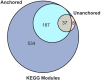
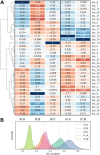
To address the question of how microbial diversity and function in the oral cavities of children relates to caries diagnosis, we surveyed the supragingival plaque biofilm microbiome in 44 juvenile twin pairs. Using shotgun sequencing, we constructed a genome encyclopedia describing the core supragingival plaque microbiome. Caries phenotypes contained statistically significant enrichments in specific genome abundances and distinct community composition profiles, including strain-level changes. Metabolic pathways that are statistically associated with caries include several sugar-associated phosphotransferase systems, antimicrobial resistance, and metal transport. Numerous closely related previously uncharacterized microbes had substantial variation in central metabolism, including the loss of biosynthetic pathways resulting in auxotrophy, changing the ecological role. We also describe the first complete Gracilibacteria genomes from the human microbiome. Caries is a microbial community metabolic disorder that cannot be described by a single etiology, and our results provide the information needed for next-generation diagnostic tools and therapeutics for caries.IMPORTANCE Oral health has substantial economic importance, with over $100 billion spent on dental care in the United States annually. The microbiome plays a critical role in oral health, yet remains poorly classified. To address the question of how microbial diversity and function in the oral cavities of children relate to caries diagnosis, we surveyed the supragingival plaque biofilm microbiome in 44 juvenile twin pairs. Using shotgun sequencing, we constructed a genome encyclopedia describing the core supragingival plaque microbiome. This unveiled several new previously uncharacterized but ubiquitous microbial lineages in the oral microbiome. Caries is a microbial community metabolic disorder that cannot be described by a single etiology, and our results provide the information needed for next-generation diagnostic tools and therapeutics for caries.
Keywords: Streptococcus; metabolism; metagenomics; microbial ecology; oral microbiology.
Copyright © 2018 Espinoza et al.
Figures



Comment in
-
Composite Metagenome-Assembled Genomes Reduce the Quality of Public Genome Repositories.mBio. 2019 Jun 4;10(3):e00725-19. doi: 10.1128/mBio.00725-19. mBio. 2019. PMID: 31164461 Free PMC article. No abstract available.
References
-
- Wall TA, Vujicic M. 2015. U.S. dental spending continues to be flat. Health Policy Institute research brief. American Dental Association, Chicago, IL.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
