Salmonella enterica Phylogeny Based on Whole-Genome Sequencing Reveals Two New Clades and Novel Patterns of Horizontally Acquired Genetic Elements
- PMID: 30482836
- PMCID: PMC6282209
- DOI: 10.1128/mBio.02303-18
Salmonella enterica Phylogeny Based on Whole-Genome Sequencing Reveals Two New Clades and Novel Patterns of Horizontally Acquired Genetic Elements
Abstract
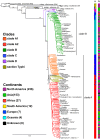
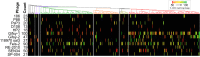
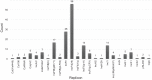
Using whole-genome sequence (WGS) data from the GenomeTrakr network, a globally distributed network of laboratories sequencing foodborne pathogens, we present a new phylogeny of Salmonella enterica comprising 445 isolates from 266 distinct serovars and originating from 52 countries. This phylogeny includes two previously unidentified S. enterica subsp. enterica clades. Serovar Typhi is shown to be nested within clade A. Our findings are supported by both phylogenetic support, based on a core genome alignment, and Bayesian approaches, based on single-nucleotide polymorphisms. Serovar assignments were refined by in silico analysis using SeqSero. More than 10% of serovars were either polyphyletic or paraphyletic. We found variable genetic content in these isolates relating to gene mobilization and virulence factors which have different distributions within clades. Gifsy-1- and Gifsy-2-like phages appear more prevalent in clade A; other viruses are more evenly distributed. Our analyses reveal IncFII is the predominant plasmid replicon in S. enterica Few core or clade-defining virulence genes are observed, and their distributions appear probabilistic in nature. Together, these patterns demonstrate that genetic exchange within S. enterica is more extensive and frequent than previously realized, which significantly alters how we view the genetic structure of the bacterial species.IMPORTANCE Rapid improvements in nucleotide sequencing access and affordability have led to a drastic increase in availability of genetic information. This information will improve the accuracy of molecular descriptions, including serovars, within S. enterica Although the concept of serovars continues to be useful, it may have more significant limitations than previously understood. Furthermore, the discrete absence or presence of specific genes can be an unstable indicator of phylogenetic identity. Whole-genome sequencing provides more rigorous tools for assessing the distributions of these genes. Our phylogenetic and genetic content analyses reveal how active genetic elements are dynamically distributed within a species, allowing us to better understand genetic reservoirs and underlying bacterial evolution.
Keywords: GenomeTrakr; Salmonella; phylogeny; plasmids; virulence; whole-genome sequencing.
Figures


References
-
- CDC. 2017. National Enteric Disease Surveillance: Salmonella annual report, 2015. https://www.cdc.gov/nationalsurveillance/salmonella-surveillance.html.
-
- Timme RE, Pettengill J, Allard MW, Strain E, Barrangou R, Wehnes C, Kessel JV, Karns J, Musser SM, Brown EW. 2013. Phylogenetic diversity of the enteric pathogen Salmonella enterica subsp. enterica inferred from genome-wide reference-free SNP characters. Genome Biol Evol 5:2109–2123. doi:10.1093/gbe/evt159. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
