The recombination mediator proteins RecFOR maintain RecA* levels for maximal DNA polymerase V Mut activity
- PMID: 30482842
- PMCID: PMC6341379
- DOI: 10.1074/jbc.RA118.005726
The recombination mediator proteins RecFOR maintain RecA* levels for maximal DNA polymerase V Mut activity
Abstract
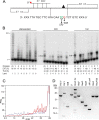
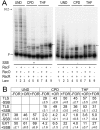
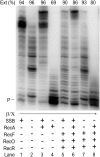
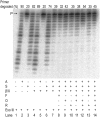
DNA template damage can potentially block DNA replication. Cells have therefore developed different strategies to repair template lesions. Activation of the bacterial lesion bypass DNA polymerase V (Pol V) requires both the cleavage of the UmuD subunit to UmuD' and the acquisition of a monomer of activated RecA recombinase, forming Pol V Mut. Both of these events are mediated by the generation of RecA* via the formation of a RecA-ssDNA filament during the SOS response. Formation of RecA* is itself modulated by competition with the ssDNA-binding protein (SSB) for binding to ssDNA. Previous observations have demonstrated that RecA filament formation on SSB-coated DNA can be favored in the presence of the recombination mediator proteins RecF, RecO, and RecR. We show here using purified proteins that in the presence of SSB and RecA, a stable RecA-ssDNA filament is not formed, although sufficient RecA* is generated to support some activation of Pol V. The presence of RecFOR increased RecA* generation and allowed Pol V to synthesize longer DNA products and to elongate from an unpaired primer terminus opposite template damage, also without the generation of a stable RecA-ssDNA filament.
Keywords: DNA damage; DNA polymerase; DNA recombination; DNA repair; RecA; lesion bypass; nucleic acid enzymology; recombination mediator protein; translesion synthesis.
© 2019 Raychaudhury and Marians.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures
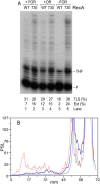
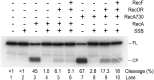
Similar articles
-
RecFOR proteins are essential for Pol V-mediated translesion synthesis and mutagenesis.EMBO J. 2006 Dec 13;25(24):5754-63. doi: 10.1038/sj.emboj.7601474. Epub 2006 Nov 30. EMBO J. 2006. PMID: 17139245 Free PMC article.
-
Conformational regulation of Escherichia coli DNA polymerase V by RecA and ATP.PLoS Genet. 2019 Feb 4;15(2):e1007956. doi: 10.1371/journal.pgen.1007956. eCollection 2019 Feb. PLoS Genet. 2019. PMID: 30716079 Free PMC article.
-
Replication restart in UV-irradiated Escherichia coli involving pols II, III, V, PriA, RecA and RecFOR proteins.Mol Microbiol. 2002 Feb;43(3):617-28. doi: 10.1046/j.1365-2958.2002.02747.x. Mol Microbiol. 2002. PMID: 11929519
-
A Comprehensive View of Translesion Synthesis in Escherichia coli.Microbiol Mol Biol Rev. 2020 Jun 17;84(3):e00002-20. doi: 10.1128/MMBR.00002-20. Print 2020 Aug 19. Microbiol Mol Biol Rev. 2020. PMID: 32554755 Free PMC article. Review.
-
Mutations for Worse or Better: Low-Fidelity DNA Synthesis by SOS DNA Polymerase V Is a Tightly Regulated Double-Edged Sword.Biochemistry. 2016 Apr 26;55(16):2309-18. doi: 10.1021/acs.biochem.6b00117. Epub 2016 Apr 12. Biochemistry. 2016. PMID: 27043933 Free PMC article. Review.
Cited by
-
The Role of the RecFOR Complex in Genome Stability.Int J Mol Sci. 2025 Jun 6;26(12):5441. doi: 10.3390/ijms26125441. Int J Mol Sci. 2025. PMID: 40564904 Free PMC article. Review.
-
RecF protein targeting to postreplication (daughter strand) gaps I: DNA binding by RecF and RecFR.Nucleic Acids Res. 2023 Jun 23;51(11):5699-5713. doi: 10.1093/nar/gkad311. Nucleic Acids Res. 2023. PMID: 37125642 Free PMC article.
-
Elucidating Recombination Mediator Function Using Biophysical Tools.Biology (Basel). 2021 Apr 1;10(4):288. doi: 10.3390/biology10040288. Biology (Basel). 2021. PMID: 33916151 Free PMC article. Review.
-
MicroRNA let-7i Inhibits Histone Lysine Demethylase KDM5B to Halt Esophageal Cancer Progression.Mol Ther Nucleic Acids. 2020 Sep 16;22:846-861. doi: 10.1016/j.omtn.2020.09.012. eCollection 2020 Dec 4. Mol Ther Nucleic Acids. 2020. PMID: 33230480 Free PMC article.
-
Single strand gap repair: The presynaptic phase plays a pivotal role in modulating lesion tolerance pathways.PLoS Genet. 2022 Jun 2;18(6):e1010238. doi: 10.1371/journal.pgen.1010238. eCollection 2022 Jun. PLoS Genet. 2022. PMID: 35653392 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

