Metabolic network-based stratification of hepatocellular carcinoma reveals three distinct tumor subtypes
- PMID: 30482855
- PMCID: PMC6294939
- DOI: 10.1073/pnas.1807305115
Metabolic network-based stratification of hepatocellular carcinoma reveals three distinct tumor subtypes
Abstract
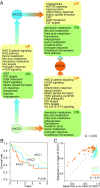
Hepatocellular carcinoma (HCC) is one of the most frequent forms of liver cancer, and effective treatment methods are limited due to tumor heterogeneity. There is a great need for comprehensive approaches to stratify HCC patients, gain biological insights into subtypes, and ultimately identify effective therapeutic targets. We stratified HCC patients and characterized each subtype using transcriptomics data, genome-scale metabolic networks and network topology/controllability analysis. This comprehensive systems-level analysis identified three distinct subtypes with substantial differences in metabolic and signaling pathways reflecting at genomic, transcriptomic, and proteomic levels. These subtypes showed large differences in clinical survival associated with altered kynurenine metabolism, WNT/β-catenin-associated lipid metabolism, and PI3K/AKT/mTOR signaling. Integrative analyses indicated that the three subtypes rely on alternative enzymes (e.g., ACSS1/ACSS2/ACSS3, PKM/PKLR, ALDOB/ALDOA, MTHFD1L/MTHFD2/MTHFD1) to catalyze the same reactions. Based on systems-level analysis, we identified 8 to 28 subtype-specific genes with pivotal roles in controlling the metabolic network and predicted that these genes may be targeted for development of treatment strategies for HCC subtypes by performing in silico analysis. To validate our predictions, we performed experiments using HepG2 cells under normoxic and hypoxic conditions and observed opposite expression patterns between genes expressed in high/moderate/low-survival tumor groups in response to hypoxia, reflecting activated hypoxic behavior in patients with poor survival. In conclusion, our analyses showed that the heterogeneous HCC tumors can be stratified using a metabolic network-driven approach, which may also be applied to other cancer types, and this stratification may have clinical implications to drive the development of precision medicine.
Keywords: biological networks; genome-scale metabolic models; hepatocellular carcinoma; personalized medicine; systems biology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Ferlay J, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. - PubMed
-
- Mardinoglu A, Boren J, Smith U, Uhlen M, Nielsen J. Systems biology in hepatology: Approaches and applications. Nat Rev Gastroenterol Hepatol. 2018;15:365–377. - PubMed
-
- O’Day E, et al. Are we there yet? How and when specific biotechnologies will improve human health. Biotechnol J. 2018:e1800195. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

