PYL8 mediates ABA perception in the root through non-cell-autonomous and ligand-stabilization-based mechanisms
- PMID: 30482863
- PMCID: PMC6294950
- DOI: 10.1073/pnas.1815410115
PYL8 mediates ABA perception in the root through non-cell-autonomous and ligand-stabilization-based mechanisms
Abstract
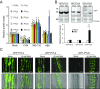
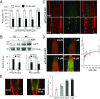
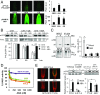
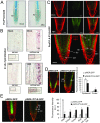
The phytohormone abscisic acid (ABA) plays a key role regulating root growth, root system architecture, and root adaptive responses, such as hydrotropism. The molecular and cellular mechanisms that regulate the action of core ABA signaling components in roots are not fully understood. ABA is perceived through receptors from the PYR/PYL/RCAR family and PP2C coreceptors. PYL8/RCAR3 plays a nonredundant role in regulating primary and lateral root growth. Here we demonstrate that ABA specifically stabilizes PYL8 compared with other ABA receptors and induces accumulation of PYL8 in root nuclei. This requires ABA perception by PYL8 and leads to diminished ubiquitination of PYL8 in roots. The ABA agonist quinabactin, which promotes root ABA signaling through dimeric receptors, fails to stabilize the monomeric receptor PYL8. Moreover, a PYL8 mutant unable to bind ABA and inhibit PP2C is not stabilized by the ligand, whereas a PYL85KR mutant is more stable than PYL8 at endogenous ABA concentrations. The PYL8 transcript was detected in the epidermis and stele of the root meristem; however, the PYL8 protein was also detected in adjacent tissues. Expression of PYL8 driven by tissue-specific promoters revealed movement to adjacent tissues. Hence both inter- and intracellular trafficking of PYL8 appears to occur in the root apical meristem. Our findings reveal a non-cell-autonomous mechanism for hormone receptors and help explain the nonredundant role of PYL8-mediated root ABA signaling.
Keywords: ABA; ABA biosensor; PYL8; non-cell-autonomous; root.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

