Evidence of a fixed internal gene constellation in influenza A viruses isolated from wild birds in Argentina (2006-2016)
- PMID: 30482896
- PMCID: PMC6258671
- DOI: 10.1038/s41426-018-0190-2
Evidence of a fixed internal gene constellation in influenza A viruses isolated from wild birds in Argentina (2006-2016)
Abstract
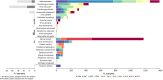
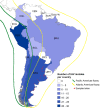
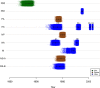
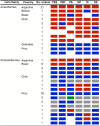
Wild aquatic birds are the major reservoir of influenza A virus. Cloacal swabs and feces samples (n = 6595) were collected from 62 bird species in Argentina from 2006 to 2016 and screened for influenza A virus. Full genome sequencing of 15 influenza isolates from 6 waterfowl species revealed subtypes combinations that were previously described in South America (H1N1, H4N2, H4N6 (n = 3), H5N3, H6N2 (n = 4), and H10N7 (n = 2)), and new ones not previously identified in the region (H4N8, H7N7 and H7N9). Notably, the internal gene segments of all 15 Argentine isolates belonged to the South American lineage, showing a divergent evolution of these viruses in the Southern Hemisphere. Time-scaled phylogenies indicated that South American gene segments diverged between ~ 30 and ~ 140 years ago from the most closely related influenza lineages, which include the avian North American (PB1, HA, NA, MP, and NS-B) and Eurasian lineage (PB2), and the equine H3N8 lineage (PA, NP, and NS-A). Phylogenetic analyses of the hemagglutinin and neuraminidase gene segments of the H4, H6, and N8 subtypes revealed recent introductions and reassortment between viruses from the Northern and Southern Hemispheres in the Americas. Remarkably and despite evidence of recent hemagglutinin and neuraminidase subtype introductions, the phylogenetic composition of internal gene constellation of these influenza A viruses has remained unchanged. Considering the extended time and the number of sampled species of the current study, and the paucity of previously available data, our results contribute to a better understanding of the ecology and evolution of influenza virus in South America.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

Similar articles
-
Genetic Characterization of Avian Influenza A (H11N9) Virus Isolated from Mandarin Ducks in South Korea in 2018.Viruses. 2020 Feb 12;12(2):203. doi: 10.3390/v12020203. Viruses. 2020. PMID: 32059510 Free PMC article.
-
Avian influenza A virus monitoring in wild birds in Bavaria: occurrence and heterogeneity of H5 and N1 encoding genes.Zoonoses Public Health. 2010 Dec;57(7-8):e184-94. doi: 10.1111/j.1863-2378.2010.01326.x. Zoonoses Public Health. 2010. PMID: 20298489
-
H6N2 reassortant avian influenza virus isolate in wild birds in Jiangxi Province, China.Virus Genes. 2024 Jun;60(3):320-324. doi: 10.1007/s11262-024-02068-5. Epub 2024 May 9. Virus Genes. 2024. PMID: 38722491
-
Epidemiology of low pathogenic avian influenza viruses in wild birds.Rev Sci Tech. 2009 Apr;28(1):49-58. doi: 10.20506/rst.28.1.1863. Rev Sci Tech. 2009. PMID: 19618618 Review.
-
Evolution and ecology of influenza A viruses.Microbiol Rev. 1992 Mar;56(1):152-79. doi: 10.1128/mr.56.1.152-179.1992. Microbiol Rev. 1992. PMID: 1579108 Free PMC article. Review.
Cited by
-
Novel Low Pathogenic Avian Influenza H6N1 in Backyard Chicken in Easter Island (Rapa Nui), Chilean Polynesia.Viruses. 2022 Mar 30;14(4):718. doi: 10.3390/v14040718. Viruses. 2022. PMID: 35458448 Free PMC article.
-
Equine-Like H3 Avian Influenza Viruses in Wild Birds, Chile.Emerg Infect Dis. 2020 Dec;26(12):2887-2898. doi: 10.3201/eid2612.202063. Emerg Infect Dis. 2020. PMID: 33219648 Free PMC article.
-
Epidemiological data of an influenza A/H5N1 outbreak in elephant seals in Argentina indicates mammal-to-mammal transmission.Nat Commun. 2024 Nov 11;15(1):9516. doi: 10.1038/s41467-024-53766-5. Nat Commun. 2024. PMID: 39528494 Free PMC article.
-
Improved detection of influenza A virus from blue-winged teals by sequencing directly from swab material.Ecol Evol. 2019 May 11;9(11):6534-6546. doi: 10.1002/ece3.5232. eCollection 2019 Jun. Ecol Evol. 2019. PMID: 31236242 Free PMC article.
-
South American H4N2 influenza A virus improved replication in chicken trachea after low number of passages.Front Vet Sci. 2023 May 31;10:1182550. doi: 10.3389/fvets.2023.1182550. eCollection 2023. Front Vet Sci. 2023. PMID: 37323837 Free PMC article.
References
MeSH terms
Substances
Grants and funding
- HHSN272201400008C/AI/NIAID NIH HHS/United States
- HHSN266200700010C/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- HHSN272201400008C/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- HHSN266200700010C/AI/NIAID NIH HHS/United States
- PICT 2008 - 0922/Consejo Nacional de Investigaciones Científicas y Técnicas (National Scientific and Technical Research Council)
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
