Regulatory Networks Involving STATs, IRFs, and NFκB in Inflammation
- PMID: 30483250
- PMCID: PMC6242948
- DOI: 10.3389/fimmu.2018.02542
Regulatory Networks Involving STATs, IRFs, and NFκB in Inflammation
Abstract
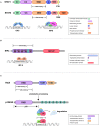
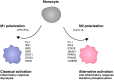
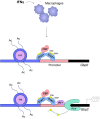
Cells engaging in inflammation undergo drastic changes of their transcriptomes. In order to tailor these alterations in gene expression to the requirements of the inflammatory process, tight and coordinate regulation of gene expression by environmental cues, microbial or danger-associated molecules or cytokines, are mandatory. The transcriptional response is set off by signal-regulated transcription factors (SRTFs) at the receiving end of pathways originating at pattern recognition- and cytokine receptors. These interact with a genome that has been set for an appropriate response by prior activity of pioneer or lineage determining transcription factors (LDTFs). The same types of transcription factors are also critical determinants of the changes in chromatin landscapes and transcriptomes that specify potential consequences of inflammation: tissue repair, training, and tolerance. Here we focus on the role of three families of SRTFs in inflammation and its sequels: signal transducers and activators of transcription (STATs), interferon regulatory factors (IRFs), and nuclear factor κB (NFκB). We describe recent findings about their interactions and about their networking with LDTFs. Our aim is to provide a snapshot of a highly dynamic research area.
Keywords: IRF; NFκB; STAT; chromatin; epigenetic; inflammation; macrophage; transcription.
Figures


Similar articles
-
Direct Inhibition of IRF-Dependent Transcriptional Regulatory Mechanisms Associated With Disease.Front Immunol. 2019 May 24;10:1176. doi: 10.3389/fimmu.2019.01176. eCollection 2019. Front Immunol. 2019. PMID: 31178872 Free PMC article. Review.
-
Developmental expression of STATs, nuclear factor-κB and inflammatory genes in the jejunum of piglets during weaning.Int Immunopharmacol. 2016 Jul;36:199-204. doi: 10.1016/j.intimp.2016.04.032. Epub 2016 May 6. Int Immunopharmacol. 2016. PMID: 27160867
-
Signal Integration of IFN-I and IFN-II With TLR4 Involves Sequential Recruitment of STAT1-Complexes and NFκB to Enhance Pro-inflammatory Transcription.Front Immunol. 2019 Jun 4;10:1253. doi: 10.3389/fimmu.2019.01253. eCollection 2019. Front Immunol. 2019. PMID: 31231385 Free PMC article.
-
The signal transducers Stat1 and Stat3 and their novel target Jmjd3 drive the expression of inflammatory genes in microglia.J Mol Med (Berl). 2014 Mar;92(3):239-54. doi: 10.1007/s00109-013-1090-5. Epub 2013 Oct 6. J Mol Med (Berl). 2014. PMID: 24097101 Free PMC article.
-
Targeted inhibition of STATs and IRFs as a potential treatment strategy in cardiovascular disease.Oncotarget. 2016 Jul 26;7(30):48788-48812. doi: 10.18632/oncotarget.9195. Oncotarget. 2016. PMID: 27166190 Free PMC article. Review.
Cited by
-
The Foot-and-Mouth Disease Virus Lb Protease Cleaves Intracellular Transcription Factors STAT1 and STAT2 to Antagonize IFN-β-Induced Signaling.J Immunol. 2023 Feb 1;210(3):283-296. doi: 10.4049/jimmunol.2101042. J Immunol. 2023. PMID: 36548461 Free PMC article.
-
STAT1 coordinates intestinal epithelial cell death during gastrointestinal infection upstream of Caspase-8.Mucosal Immunol. 2022 Jan;15(1):130-142. doi: 10.1038/s41385-021-00450-2. Epub 2021 Sep 8. Mucosal Immunol. 2022. PMID: 34497340 Free PMC article.
-
Microglia Phenotypes in Aging and Neurodegenerative Diseases.Cells. 2022 Jun 30;11(13):2091. doi: 10.3390/cells11132091. Cells. 2022. PMID: 35805174 Free PMC article. Review.
-
Immunomodulatory Nanoparticles Mitigate Macrophage Inflammation via Inhibition of PAMP Interactions and Lactate-Mediated Functional Reprogramming of NF-κB and p38 MAPK.Pharmaceutics. 2021 Nov 2;13(11):1841. doi: 10.3390/pharmaceutics13111841. Pharmaceutics. 2021. PMID: 34834256 Free PMC article.
-
The Yin and Yang of Type I IFNs in Cancer Promotion and Immune Activation.Biology (Basel). 2021 Sep 1;10(9):856. doi: 10.3390/biology10090856. Biology (Basel). 2021. PMID: 34571733 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

