Echovirus 30 and coxsackievirus A9 infection among young neonates with sepsis in Iran
- PMID: 30483379
- PMCID: PMC6243150
Echovirus 30 and coxsackievirus A9 infection among young neonates with sepsis in Iran
Abstract
Background and objectives: Human enteroviruses (EV) are the most common causes of neonatal sepsis-like disease. The frequencies of EV including coxsackievirus A, coxsackievirus B and Echovirus serotypes have been studied in young infants (younger than three months) with sepsis. So far, the role of enteroviruses among neonates with sepsis was not determined in Ahvaz, Iran. Therefore, this study was aimed to evaluate the frequency of EV among hospitalized young infants with clinical signs and symptoms of sepsis in Ahvaz.
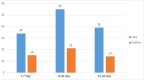
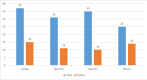
Materials and methods: Blood specimens from 128 neonates (younger than 90 days), including 56 (43.75%) girls and 72 (56.25%) boys, were collected from hospitalized neonates with clinical signs and symptoms of sepsis-like symptoms. All blood samples were negative for bacterial culture. RNA was extracted from all sera and tested for detection of 5'UTR (Untranslated Region) of the EV by RT-PCR. To determine specific strains of EV, positive 5'UTR samples were further tested for detection of the VP1 region of EV by RT-PCR.
Results: Overall, 50/128 (39.06%) specimens, including 24 (48%) girls and 26 (52%) boys, were positive for EV. 21/50 (42%) specimens were positive for the VP1 region. Randomly, 8 positive VP1 were selected and sequenced. Analysis of sequencing data showed 7/21 (33.33%) samples were positive for Echovirus 30 and 1/21 (4.76%) samples were positive for CVA9.
Conclusion: The results of this survey indicate high prevalence of 39.06% of EV among young neonates with sepsis. A high prevalence of 33.3% Echoviruses 30 and a low rate of 4.76% coxsackievirus A9 infection has been observed in neonatal patients with viral sepsis. This outbreak is probably one of the first Enterovirus outbreaks to be reported in Ahvaz, Iran. The results of this survey will help to minimize unneeded use of antimicrobial drugs and reduce unnecessary hospitalization.
Keywords: Coxsackievirus; Echovirus; Enteroviruses; Sepsis.
Figures


References
-
- Knowles NJ., Hovi T., Hyypiä T., King AMQ., Lindberg AM., Pallansch MA., Palmenberg AC., Skern T., Stanway G., Yamashita T., Zell R. Picornaviridae. In: King AMQ., Adams MJ., Carstens EB., Lefkowitz EJ., editors. Virus taxonomy: classification and nomenclature of viruses: ninth report of the international committee on Taxonomy of viruses. Elsevier; San Diego: 2011. p. 855–880.
-
- Knowles NJ, Hovi T, Hyypi T, King AM, Lindberg AM, Pallansch MA, et al. Picornaviridae. In: King AM, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Virus Taxonomy: Classification and Nomenclature of Viruses. Ninth Report of the International Committee on Taxonomy of Viruses. San Diego: Elsevier; 2012. p. 855–80.
-
- Ahmad S, Dalwai A, Al-Nakib W. Frequency of enterovirus detection in blood samples of neonates admitted to hospital with sepsis-like illness in Kuwait. J Med Virol 2013;85:1280–1285. - PubMed
-
- Harvala H, Calvert J, Van Nguyen D, Clasper L, Gadsby N, Molyneaux P, et al. Comparison of diagnostic clinical samples and environmental sampling for enterovirus and parechovirus surveillance in Scotland, 2010 to 2012. Euro Surveill 2014;19(15):20772. - PubMed
-
- Maguire HC, Atkinson P, Sharland M, Bendig J. Enterovirus infections in England and Wales: laboratory surveillance data: 1975 to 1994. Commun Dis Public Health 1999;2:122–125. - PubMed
LinkOut - more resources
Full Text Sources
