Computational Modeling and Reverse Engineering to Reveal Dominant Regulatory Interactions Controlling Osteochondral Differentiation: Potential for Regenerative Medicine
- PMID: 30483498
- PMCID: PMC6243751
- DOI: 10.3389/fbioe.2018.00165
Computational Modeling and Reverse Engineering to Reveal Dominant Regulatory Interactions Controlling Osteochondral Differentiation: Potential for Regenerative Medicine
Abstract
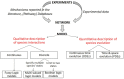
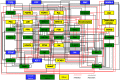
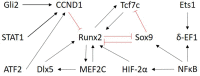
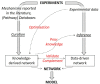
The specialization of cartilage cells, or chondrogenic differentiation, is an intricate and meticulously regulated process that plays a vital role in both bone formation and cartilage regeneration. Understanding the molecular regulation of this process might help to identify key regulatory factors that can serve as potential therapeutic targets, or that might improve the development of qualitative and robust skeletal tissue engineering approaches. However, each gene involved in this process is influenced by a myriad of feedback mechanisms that keep its expression in a desirable range, making the prediction of what will happen if one of these genes defaults or is targeted with drugs, challenging. Computer modeling provides a tool to simulate this intricate interplay from a network perspective. This paper aims to give an overview of the current methodologies employed to analyze cell differentiation in the context of skeletal tissue engineering in general and osteochondral differentiation in particular. In network modeling, a network can either be derived from mechanisms and pathways that have been reported in the literature (knowledge-based approach) or it can be inferred directly from the data (data-driven approach). Combinatory approaches allow further optimization of the network. Once a network is established, several modeling technologies are available to interpret dynamically the relationships that have been put forward in the network graph (implication of the activation or inhibition of certain pathways on the evolution of the system over time) and to simulate the possible outcomes of the established network such as a given cell state. This review provides for each of the aforementioned steps (building, optimizing, and modeling the network) a brief theoretical perspective, followed by a concise overview of published works, focusing solely on applications related to cell fate decisions, cartilage differentiation and growth plate biology. Particular attention is paid to an in-house developed example of gene regulatory network modeling of growth plate chondrocyte differentiation as all the aforementioned steps can be illustrated. In summary, this paper discusses and explores a series of tools that form a first step toward a rigorous and systems-level modeling of osteochondral differentiation in the context of regenerative medicine.
Keywords: chondrocyte; differentiation; gene regulatory network; in silico modeling; network inference; regenerative medicine.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources

