The PII-NAGK-PipX-NtcA Regulatory Axis of Cyanobacteria: A Tale of Changing Partners, Allosteric Effectors and Non-covalent Interactions
- PMID: 30483512
- PMCID: PMC6243067
- DOI: 10.3389/fmolb.2018.00091
The PII-NAGK-PipX-NtcA Regulatory Axis of Cyanobacteria: A Tale of Changing Partners, Allosteric Effectors and Non-covalent Interactions
Abstract
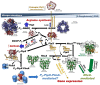
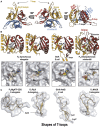
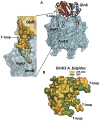
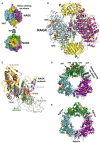
PII, a homotrimeric very ancient and highly widespread (bacteria, archaea, plants) key sensor-transducer protein, conveys signals of abundance or poorness of carbon, energy and usable nitrogen, converting these signals into changes in the activities of channels, enzymes, or of gene expression. PII sensing is mediated by the PII allosteric effectors ATP, ADP (and, in some organisms, AMP), 2-oxoglutarate (2OG; it reflects carbon abundance and nitrogen scarcity) and, in many plants, L-glutamine. Cyanobacteria have been crucial for clarification of the structural bases of PII function and regulation. They are the subject of this review because the information gathered on them provides an overall structure-based view of a PII regulatory network. Studies on these organisms yielded a first structure of a PII complex with an enzyme, (N-acetyl-Lglutamate kinase, NAGK), deciphering how PII can cause enzyme activation, and how it promotes nitrogen stockpiling as arginine in cyanobacteria and plants. They have also revealed the first clear-cut mechanism by which PII can control gene expression. A small adaptor protein, PipX, is sequestered by PII when nitrogen is abundant and is released when is scarce, swapping partner by binding to the 2OG-activated transcriptional regulator NtcA, co-activating it. The structures of PII-NAGK, PII-PipX, PipX alone, of NtcA in inactive and 2OG-activated forms and as NtcA-2OG-PipX complex, explain structurally PII regulatory functions and reveal the changing shapes and interactions of the T-loops of PII depending on the partner and on the allosteric effectors bound to PII. Cyanobacterial studies have also revealed that in the PII-PipX complex PipX binds an additional transcriptional factor, PlmA, thus possibly expanding PipX roles beyond NtcA-dependency. Further exploration of these roles has revealed a functional interaction of PipX with PipY, a pyridoxal-phosphate (PLP) protein involved in PLP homeostasis whose mutations in the human ortholog cause epilepsy. Knowledge of cellular levels of the different components of this PII-PipX regulatory network and of KD values for some of the complexes provides the basic background for gross modeling of the system at high and low nitrogen abundance. The cyanobacterial network can guide searches for analogous components in other organisms, particularly of PipX functional analogs.
Keywords: NtcA structure and complexes; PII complexes; PipX complexes; PlmA; gene expression regulation; nitrogen regulation; protein structure; signaling.
Figures
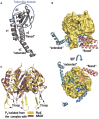
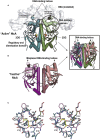
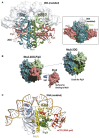

References
-
- Berg O. G., von Hippel P. H. (1988). Selection of DNA binding sites by regulatory proteins. II. The binding specificity of cyclic AMP receptor protein to recognition sites. J. Mol. Biol. 200, 709–723. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

