Conventional and emerging roles of the energy sensor Snf1/AMPK in Saccharomyces cerevisiae
- PMID: 30483520
- PMCID: PMC6244292
- DOI: 10.15698/mic2018.11.655
Conventional and emerging roles of the energy sensor Snf1/AMPK in Saccharomyces cerevisiae
Abstract
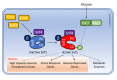
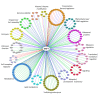
All proliferating cells need to match metabolism, growth and cell cycle progression with nutrient availability to guarantee cell viability in spite of a changing environment. In yeast, a signaling pathway centered on the effector kinase Snf1 is required to adapt to nutrient limitation and to utilize alternative carbon sources, such as sucrose and ethanol. Snf1 shares evolutionary conserved functions with the AMP-activated Kinase (AMPK) in higher eukaryotes which, activated by energy depletion, stimulates catabolic processes and, at the same time, inhibits anabolism. Although the yeast Snf1 is best known for its role in responding to a number of stress factors, in addition to glucose limitation, new unconventional roles of Snf1 have recently emerged, even in glucose repressing and unstressed conditions. Here, we review and integrate available data on conventional and non-conventional functions of Snf1 to better understand the complexity of cellular physiology which controls energy homeostasis.
Keywords: DNA damage; aging; budding yeast; cell cycle; endocytosis; glucose repression; metabolism; signaling; stress response; transcription.
Conflict of interest statement
Conflict of interest: The authors have no conflicts of interest to declare.
Figures
References
Publication types
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
