MicroRNA-30e inhibits adhesion, migration, invasion and cell cycle progression of prostate cancer cells via inhibition of the activation of the MAPK signaling pathway by downregulating CHRM3
- PMID: 30483762
- PMCID: PMC6317654
- DOI: 10.3892/ijo.2018.4647
MicroRNA-30e inhibits adhesion, migration, invasion and cell cycle progression of prostate cancer cells via inhibition of the activation of the MAPK signaling pathway by downregulating CHRM3
Retraction in
-
[Retracted] MicroRNA‑30e inhibits adhesion, migration, invasion and cell cycle progression of prostate cancer cells via inhibition of the activation of the MAPK signaling pathway by downregulating CHRM3.Int J Oncol. 2024 Sep;65(3):82. doi: 10.3892/ijo.2024.5670. Epub 2024 Jul 19. Int J Oncol. 2024. PMID: 39027991 Free PMC article.
Abstract
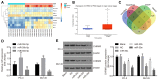
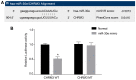
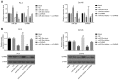
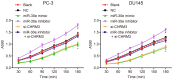
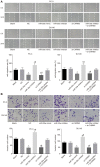
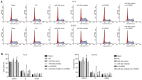
Prostate cancer (PCa) testing is currently based on measurement of serum prostate‑specific antigen levels and digital rectal examination, which are limited by a low predictive value and the adverse effects associated with overdiagnosis and overtreatment. Recent studies have reported that the abnormal expression of microRNAs (miRNAs) is associated with the mechanism underlying the development of PCa. Thus, the aim of the present study was to investigate the effects of miR‑30e and its target gene, M3 muscarinic acetylcholine receptor (CHRM3), on the adhesion, migration, invasion and cell cycle distribution of PCa cells via the mitogen‑activated protein kinase (MAPK) signaling pathway. The differentially expressed genes were screened in the Gene Expression Omnibus database from a gene expression microarray (GSE55945) of PCa. PCa tissues and adjacent tissues were collected from patients with PCa. The PC‑3 and DU145 human PCa cell lines were treated with activator, inhibitor and siRNAs. The effects of miR‑30e on cell adhesion, migration, invasion and cell cycle distribution with the involvement of CHRM3 and the MAPK signaling pathway were investigated. The bioinformatics results demonstrated that the CHRM3 gene and the MAKP signaling pathway were involved in the progression of PCa, and has‑miR‑30e was selected for further study. The levels of miR‑30e were significantly downregulated, while the levels of CHRM3 were obviously upregulated in PCa. CHRM3 was verified as a target gene of miR‑30e. Upregulation of miR‑30e and downregulation of CHRM3 decreased the levels of p‑P38, p‑extracellular signal‑regulated kinase, p‑c‑Jun N‑terminal kinase, p‑c‑fos and p‑c‑JUN, cell adhesion, migration and invasion ability, and the number of cells in the S phase, while they increased the number of cells in the G0 and G1 phases. The findings of the present study suggest that miR‑30e inhibited the adhesion, migration, invasion and cell cycle entry of PCa cells by suppressing the activation of the MAPK signaling pathway and inhibiting CHRM3 expression. Thus, miR‑30e may serve as a candidate target for the treatment of PCa.
Keywords: microRNA-30e; M3 muscarinic acetylcholine receptor; mitogen-activated protein kinase signaling pathway; prostate cancer cells; adhesion; migration; invasion; cell cycle.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

