Overexpression of MUC1 predicts poor prognosis in patients with breast cancer
- PMID: 30483806
- PMCID: PMC6313072
- DOI: 10.3892/or.2018.6887
Overexpression of MUC1 predicts poor prognosis in patients with breast cancer
Abstract
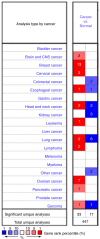
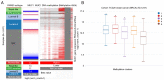
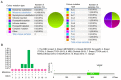
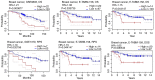
Breast cancer is the most commonly diagnosed cancer in females; thus, there is an urgent requirement to identify precise biomarkers for the diagnosis and treatment of the disease. Mucin 1 (MUC1) is a glycoprotein that has been demonstrated to be involved in the metastasis and invasion of multiple tumor types. Bioinformatics analyses were conducted to indicate the prognostic value of MUC1 in breast cancer. Additionally, the expression level of MUC1 was assessed using Oncomine analysis. Furthermore, PrognoScan was used to analyze the prognostic value of MUC1 in breast cancer. Mutations of MUC1 were analyzed by the Catalogue of Somatic Mutations in Cancer and cBioPortal databases. In addition, University of California, Santa Cruz (UCSC) was used to examine the methylation status of MUC1. Co‑expression of MUC1 mRNA was detected with the cBioPortal, UCSC and Breast Cancer Gene‑Expression Miner v4.0 datasets. The results demonstrated that MCU1 is frequently overexpressed in breast cancer and is negatively associated with CpG sites. Furthermore, pooled data indicated that abnormally high expression of MUC1 indicates poor prognosis. Additionally, upregulation of MUC1 expression is associated with estrogen receptor‑ and progesterone receptor‑positive disease, aging and increased Scarff, Bloom and Richardson grade, but is not associated with triple‑negative and basal‑like status. Subsequent data mining across multiple large databases demonstrated a positive association between MUC1 mRNA expression and cyclic AMP‑responsive element‑binding protein 3‑like 4 (CREB3L4) in breast cancer tissues. The present data indicated that the overexpression of MUC1 indicates a poor prognosis in patients with breast cancer and is associated with MUC1 promoter methylation status. Additionally, MUC1 positively correlated with CREB3L4 and may serve as a potential prognostic factor and therapy target for breast cancer.
Figures




Similar articles
-
Mining Prognostic Significance of MEG3 in Human Breast Cancer Using Bioinformatics Analysis.Cell Physiol Biochem. 2018;50(1):41-51. doi: 10.1159/000493956. Epub 2018 Oct 2. Cell Physiol Biochem. 2018. PMID: 30278461
-
Evaluation of Mucin-1 protein and mRNA expression as prognostic and predictive markers after neoadjuvant chemotherapy for breast cancer.Ann Oncol. 2013 Sep;24(9):2316-24. doi: 10.1093/annonc/mdt162. Epub 2013 May 9. Ann Oncol. 2013. PMID: 23661292 Clinical Trial.
-
Cell division cycle proteinising prognostic biomarker of breast cancer.Biosci Rep. 2020 May 29;40(5):BSR20191227. doi: 10.1042/BSR20191227. Biosci Rep. 2020. PMID: 32285914 Free PMC article.
-
MUC1 induces tamoxifen resistance in estrogen receptor-positive breast cancer.Expert Rev Anticancer Ther. 2017 Jul;17(7):607-613. doi: 10.1080/14737140.2017.1340837. Epub 2017 Jun 21. Expert Rev Anticancer Ther. 2017. PMID: 28597750 Review.
-
MUC1: Structure, Function, and Clinic Application in Epithelial Cancers.Int J Mol Sci. 2021 Jun 18;22(12):6567. doi: 10.3390/ijms22126567. Int J Mol Sci. 2021. PMID: 34207342 Free PMC article. Review.
Cited by
-
AIbZIP/CREB3L4 Promotes Cell Proliferation via the SKP2-p27 Axis in Luminal Androgen Receptor Subtype Triple-Negative Breast Cancer.Mol Cancer Res. 2024 Apr 2;22(4):373-385. doi: 10.1158/1541-7786.MCR-23-0629. Mol Cancer Res. 2024. PMID: 38236913 Free PMC article.
-
The Complex Interaction between the Tumor Micro-Environment and Immune Checkpoints in Breast Cancer.Cancers (Basel). 2019 Aug 19;11(8):1205. doi: 10.3390/cancers11081205. Cancers (Basel). 2019. PMID: 31430935 Free PMC article.
-
Chimeric Antigen Receptor (CAR)-T Cell Immunotherapy Against Thoracic Malignancies: Challenges and Opportunities.Front Immunol. 2022 Jul 14;13:871661. doi: 10.3389/fimmu.2022.871661. eCollection 2022. Front Immunol. 2022. PMID: 35911706 Free PMC article.
-
Amphicrine Medullary Thyroid Carcinoma - a Case-Based Review Expanding on Its MUC Expression Profile.Endocr Pathol. 2022 Sep;33(3):378-387. doi: 10.1007/s12022-022-09725-1. Epub 2022 Jun 23. Endocr Pathol. 2022. PMID: 35733030 Review.
-
Mucin 1 promotes tumor progression through activating WNT/β-catenin signaling pathway in intrahepatic cholangiocarcinoma.J Cancer. 2021 Oct 3;12(23):6937-6947. doi: 10.7150/jca.63235. eCollection 2021. J Cancer. 2021. PMID: 34729096 Free PMC article.
References
-
- Qiu N, He Y, Zhang S, Hu X, Chen M, Li H. Cullin 7 is a predictor of poor prognosis in breast cancer patients and is involved in the proliferation and invasion of breast cancer cells by regulating the cell cycle and microtubule stability. Oncol Rep. 2018;39:603–610. - PubMed
-
- Zhang X, Rice M, Tworoger SS, Rosner BA, Eliassen AH, Tamimi RM, Joshi AD, Lindstrom S, Qian J, Colditz GA, et al. Addition of a polygenic risk score, mammographic density, and endogenous hormones to existing breast cancer risk prediction models: A nested case-control study. PLoS Med. 2018;15:e1002644. doi: 10.1371/journal.pmed.1002644. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

