Three-dimensional descriptors for aminergic GPCRs: dependence on docking conformation and crystal structure
- PMID: 30484023
- PMCID: PMC6682580
- DOI: 10.1007/s11030-018-9894-4
Three-dimensional descriptors for aminergic GPCRs: dependence on docking conformation and crystal structure
Abstract
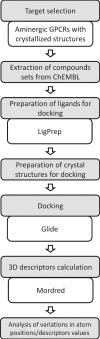
Three-dimensional descriptors are often used to search for new biologically active compounds, in both ligand- and structure-based approaches, capturing the spatial orientation of molecules. They frequently constitute an input for machine learning-based predictions of compound activity or quantitative structure-activity relationship modeling; however, the distribution of their values and the accuracy of depicting compound orientations might have an impact on the power of the obtained predictive models. In this study, we analyzed the distribution of three-dimensional descriptors calculated for docking poses of active and inactive compounds for all aminergic G protein-coupled receptors with available crystal structures, focusing on the variation in conformations for different receptors and crystals. We demonstrated that the consistency in compound orientation in the binding site is rather not correlated with the affinity itself, but is more influenced by other factors, such as the number of rotatable bonds and crystal structure used for docking studies. The visualizations of the descriptors distributions were prepared and made available online at http://chem.gmum.net/vischem_stability , which enables the investigation of chemical structures referring to particular data points depicted in the figures. Moreover, the performed analysis can assist in choosing crystal structure for docking studies, helping in selection of conditions providing the best discrimination between active and inactive compounds in machine learning-based experiments.
Keywords: Aminergic GPCRs; Crystal structure; Docking; Machine learning; Three-dimensional descriptors.
Figures


References
-
- Breda A, Basso LA, Santos DS, de Azevedo WF., Jr Virtual screening of drugs: score functions, docking, and drug design. Curr Comput Aided Drug Des. 2008;4:265–272. doi: 10.2174/157340908786786047. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

