Nature and nurture: confluence of pathway determinism with metabolic and chemical serendipity diversifies Monascus azaphilone pigments
- PMID: 30484470
- PMCID: PMC6470053
- DOI: 10.1039/c8np00060c
Nature and nurture: confluence of pathway determinism with metabolic and chemical serendipity diversifies Monascus azaphilone pigments
Abstract
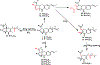
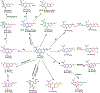
Covering: up to June 2018 Understanding the biosynthetic mechanisms that generate the astounding structural complexity and variety of fungal secondary metabolites (FSMs) remains a challenge. As an example, the biogenesis of the Monascus azaphilone pigments (MonAzPs) has remained obscure until recently despite the significant medical potential of these metabolites and their long history of widespread use as food colorants. However, a considerable progress has been made in recent years towards the elucidation of MonAzPs biosynthesis in various fungi. In this highlight, we correlate a unified biosynthetic pathway with the diverse structures of the 111 MonAzPs congeners reported until June 2018. We also discuss the origins of structural diversity amongst MonAzPs analogues and summarize new research directions towards exploring novel MonAzPs. The case of MonAzPs illuminates the various ways that FSMs metabolic complexity emerges by the interplay of biosynthetic pathway determinism with metabolic and chemical serendipity.
Figures


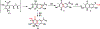


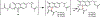



References
-
- Chen W, He Y, Zhou Y, Shao Y, Feng Y, Li M and Chen F, Compr. Rev. Food Sci, 2015, 14, 555–567.
-
- Mapari SAS, Thrane U and Meyer AS, Trends Biotechnol, 2010, 28, 300–307. - PubMed
-
- National Food Safety Standards GB1886.181–2016, 2016, 1–5.
-
- Commission Regulation (EU) No 212/2014, 2014, 3–4.
-
- Gupta RC, Lasher MA, Mukherjee IRM, Srivastava A and Lall R, in Reproductive and Developmental Toxicology (Second Edition), ed. Gupta RC, Academic Press, 2017, DOI: 10.1016/B978-0-12-804239-7.00048-2, pp. 945–962. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

